all_times <- list() # store the time for each chunk
knitr::knit_hooks$set(time_it = local({
now <- NULL
function(before, options) {
if (before) {
now <<- Sys.time()
} else {
res <- difftime(Sys.time(), now, units = "secs")
all_times[[options$label]] <<- res
}
}
}))
knitr::opts_chunk$set(
tidy = TRUE,
tidy.opts = list(width.cutoff = 95),
message = FALSE,
warning = FALSE,
fig.width = 10,
time_it = TRUE,
error = TRUE
)
load data from h5ad
t0_CreateObject <- system.time({
mat <- open_matrix_dir("../data/mouse_1M_neurons_counts")[, 1:1e+05]
mat <- Azimuth::ConvertEnsembleToSymbol(mat = mat, species = "mouse")
options(Seurat.object.assay.version = "v5", Seurat.object.assay.calcn = T)
obj <- CreateSeuratObject(counts = mat)
})
Sketch assay clustering
## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 5000
## Number of edges: 187688
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.9013
## Number of communities: 21
## Elapsed time: 0 seconds
obj <- RunUMAP(obj, dims = 1:50, return.model = T)
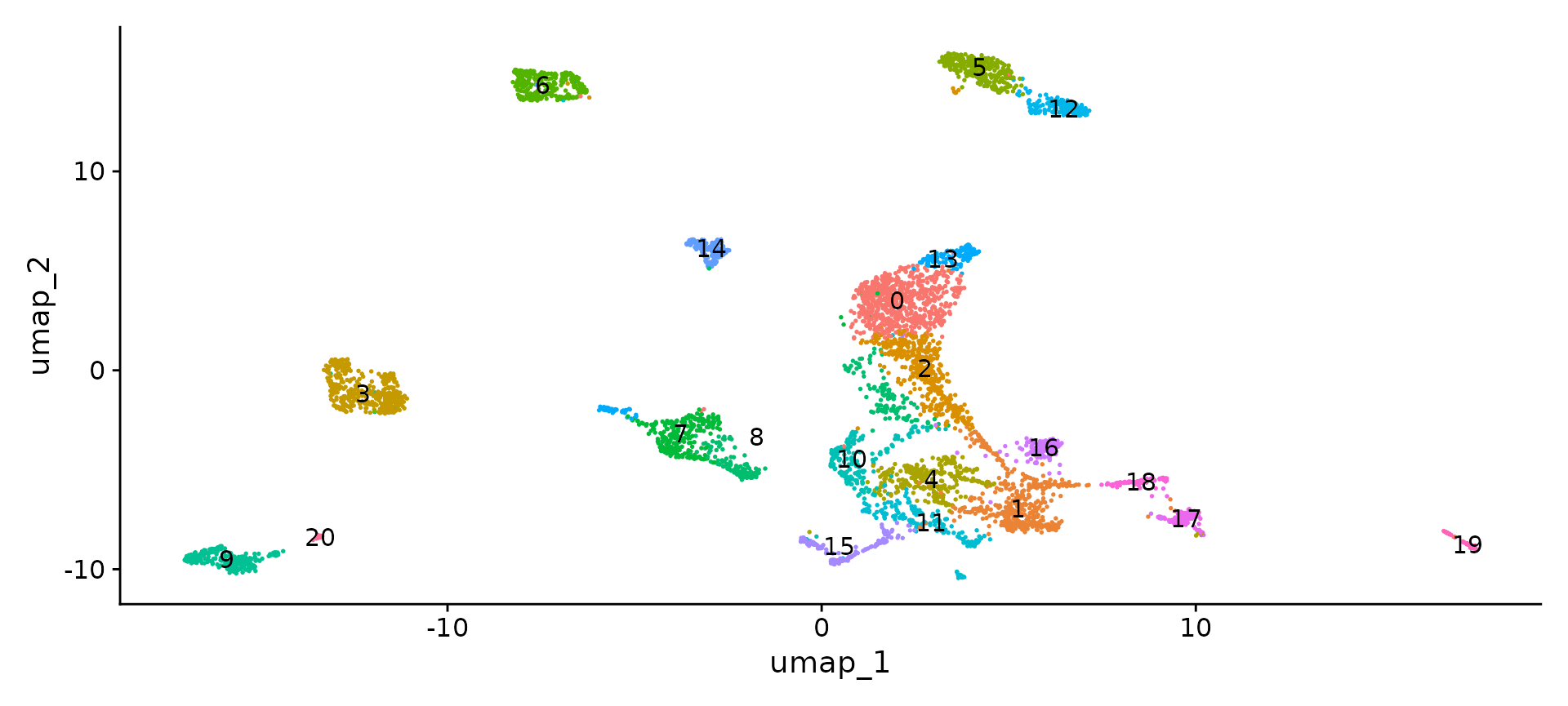
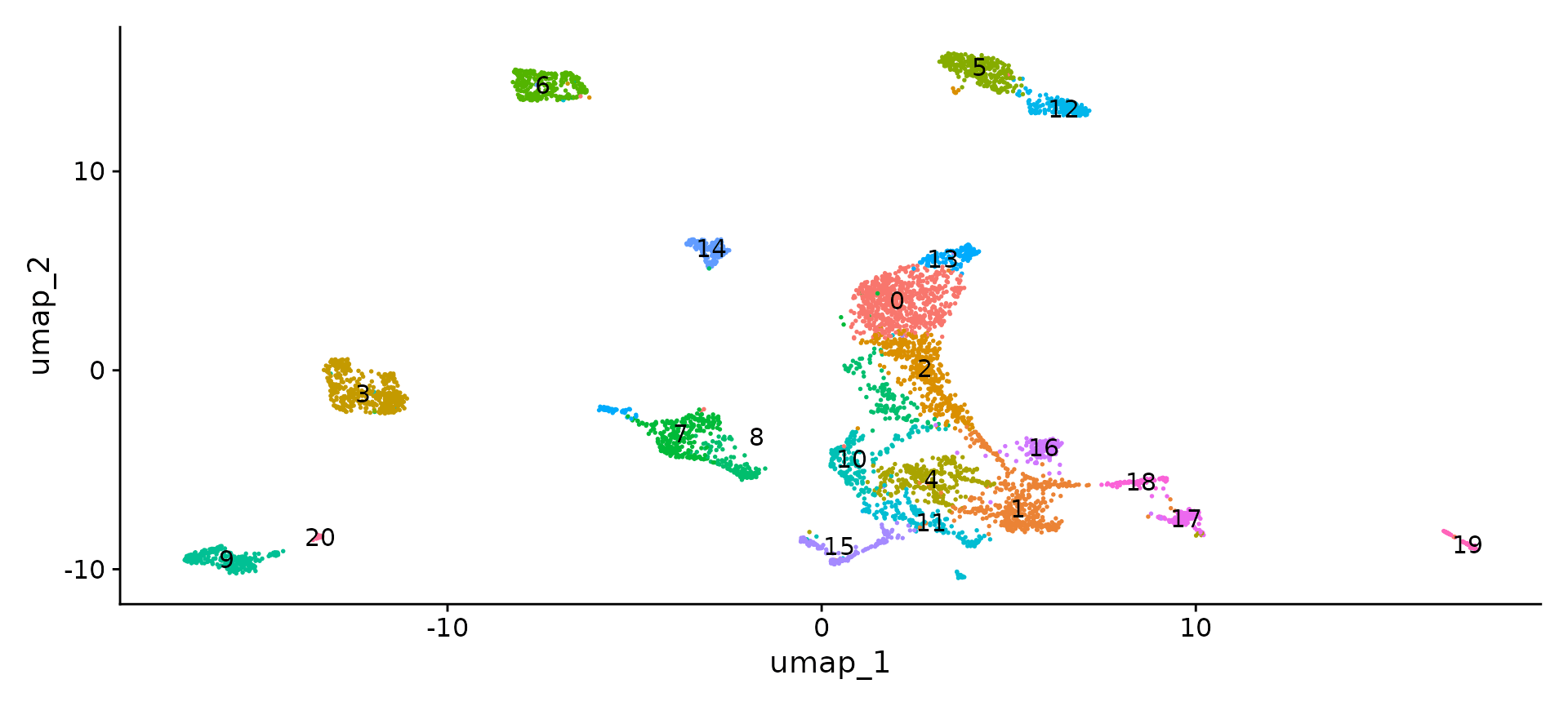
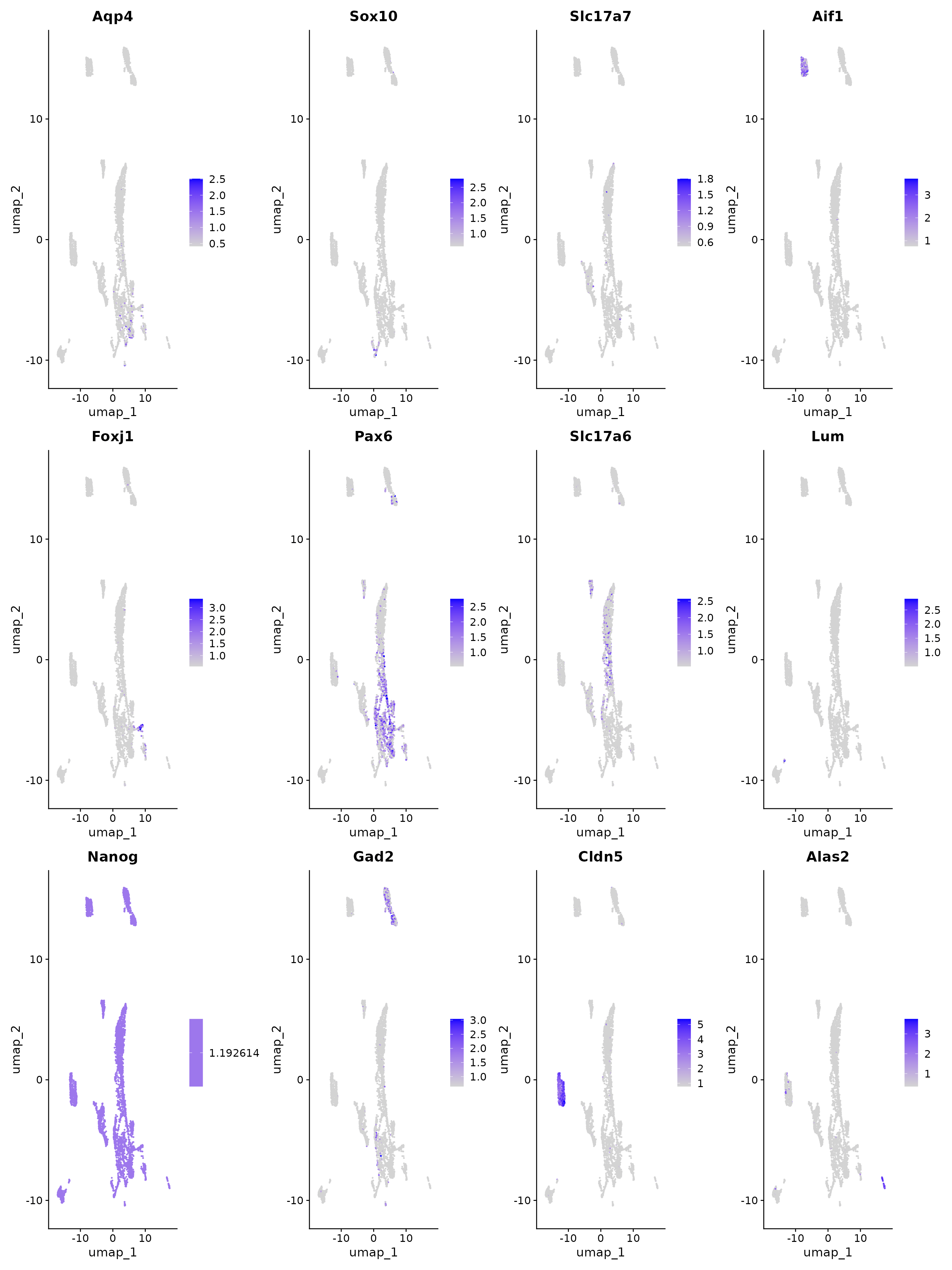
features.set <- c("Aqp4", "Sox10", "Slc17a7", "Aif1", "Foxj1", "Pax6", "Slc17a6", "Lum", "Nanog",
"Gad2", "Foxj1", "Cldn5", "Alas2")
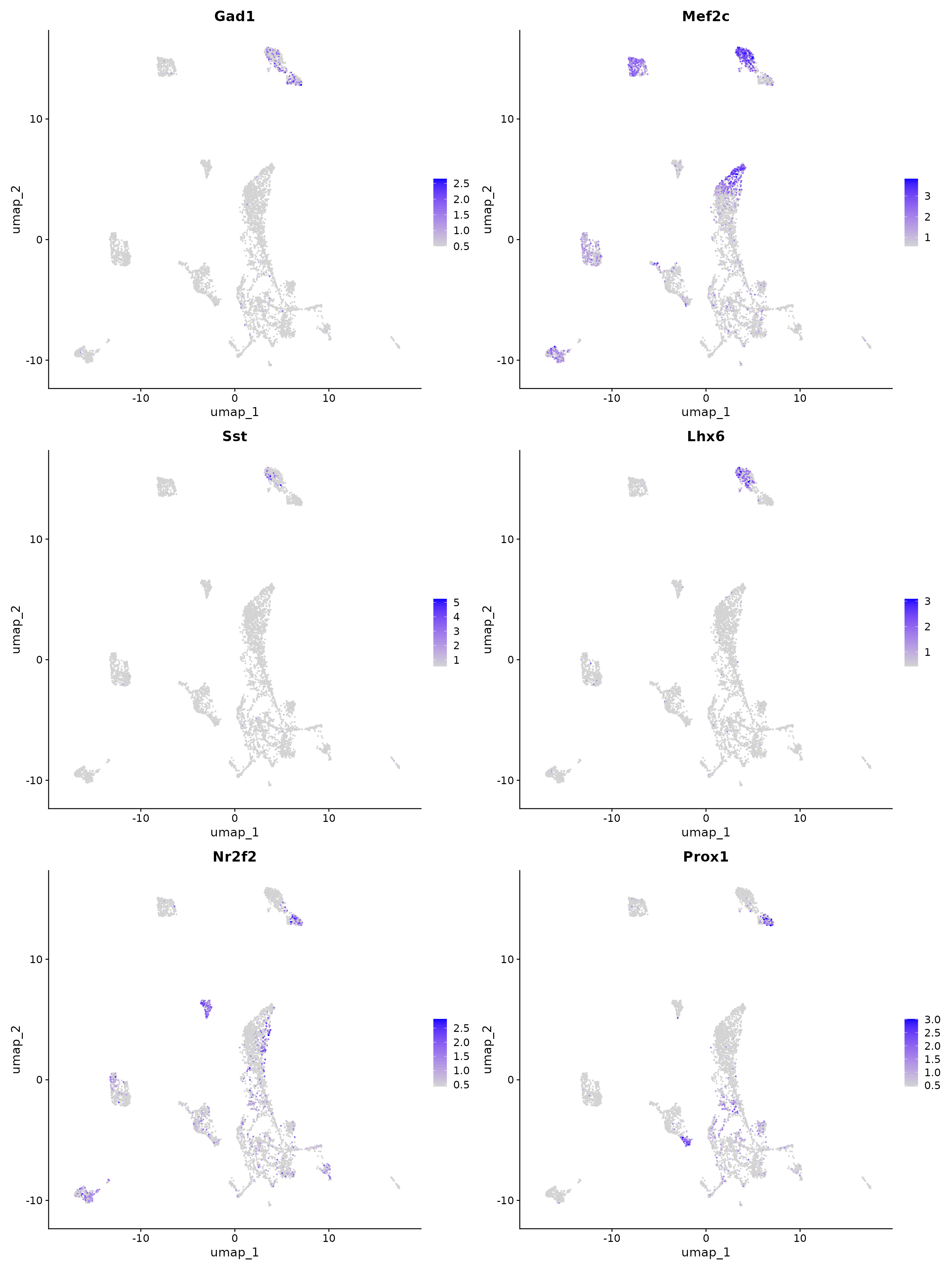
features.gaba.set <- c("Gad1", "Mef2c", "Sst", "Lhx6", "Nr2f2", "Prox1")
DefaultAssay(obj) <- "sketch"
FeaturePlot(obj, reduction = "umap", features = features.set, max.cutoff = "q99", min.cutoff = "q1")
FeaturePlot(obj, reduction = "umap", features = features.gaba.set, max.cutoff = "q99", min.cutoff = "q1")
Project full cells to PCA from sketch assay
t3_ProjectEmbedding <- system.time({
ref.emb <- ProjectCellEmbeddings(query = obj, reference = obj, query.assay = "RNA", reference.assay = "sketch",
reduction = "pca")
obj[["pca.orig"]] <- CreateDimReducObject(embeddings = ref.emb, assay = "RNA")
DefaultAssay(obj) <- "RNA"
})
Transfer labels and umap from sketch to full data
t4_transferLabel <- system.time({
options(future.globals.maxSize = 1e+09)
obj <- TransferSketchLabels(object = obj, atoms = "sketch", reduction = "pca.orig", dims = 1:50,
refdata = list(cluster_full = "sketch_snn_res.0.8"), reduction.model = "umap")
})
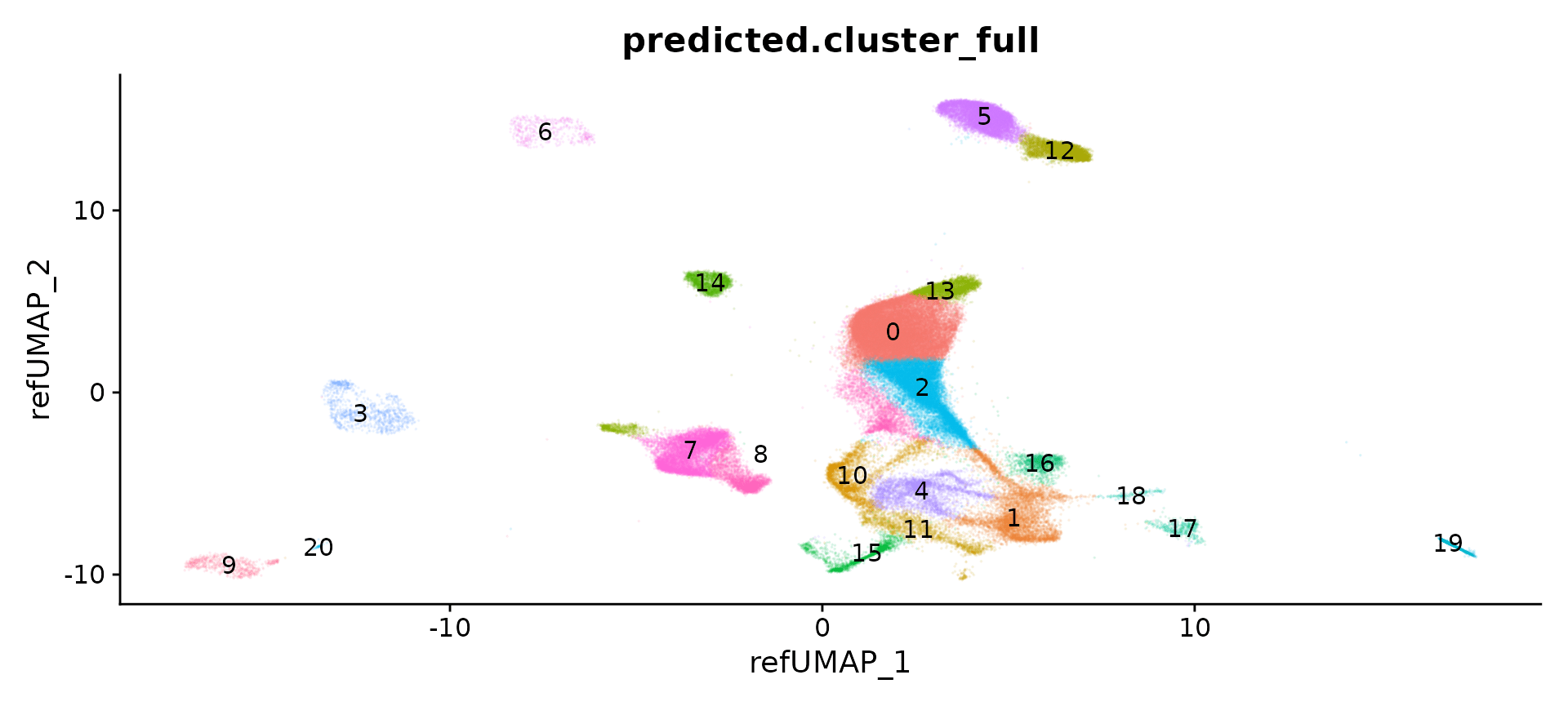
obj[["pca.nn"]] <- Seurat:::NNHelper(data = obj[["pca.orig"]]@cell.embeddings[, 1:50], k = 30, method = "hnsw",
metric = "cosine", n_threads = 10)
obj <- RunUMAP(obj, nn.name = "pca.nn", reduction.name = "umap.orig", reduction.key = "Uo_")
DimPlot(obj, label = T, reduction = "umap.orig", group.by = "predicted.cluster_full", alpha = 0.1) +
NoLegend()
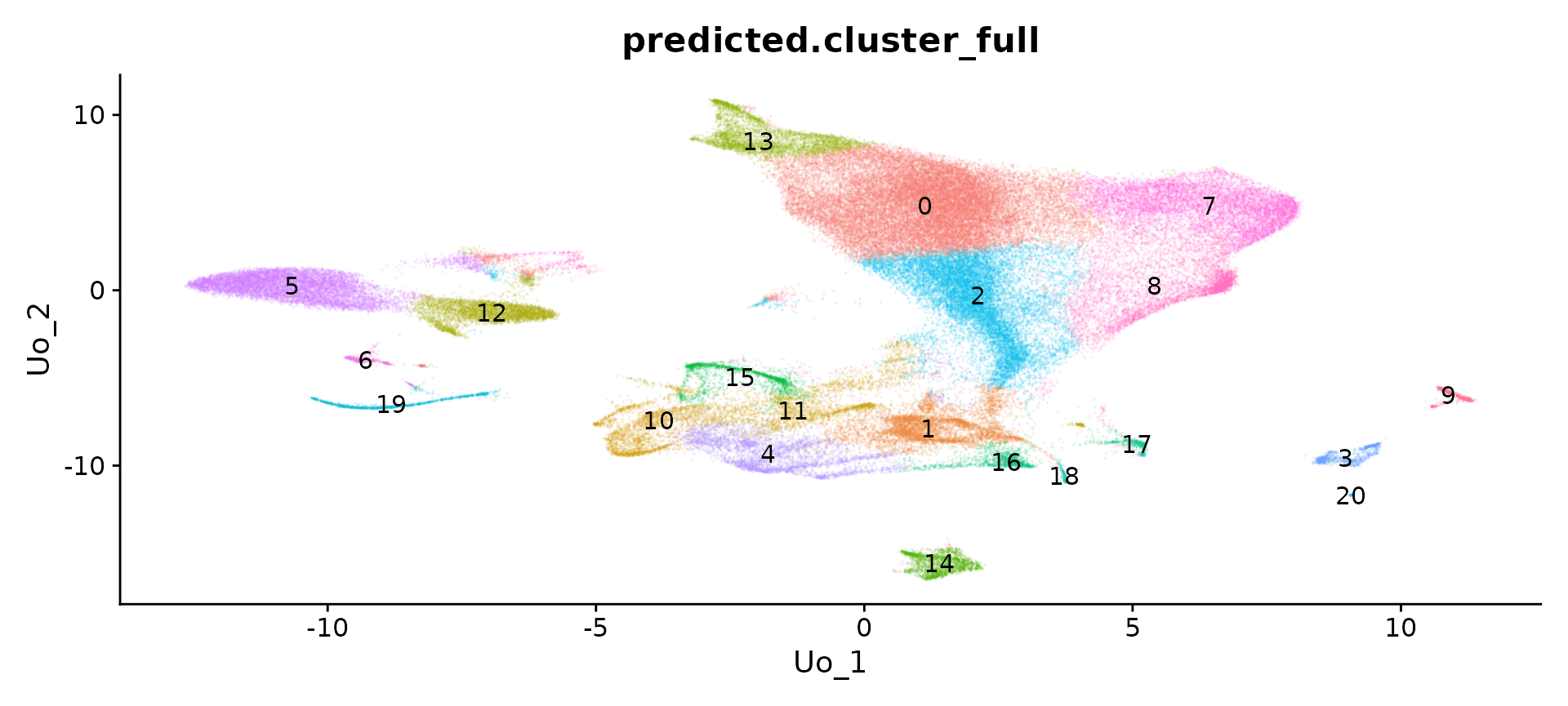
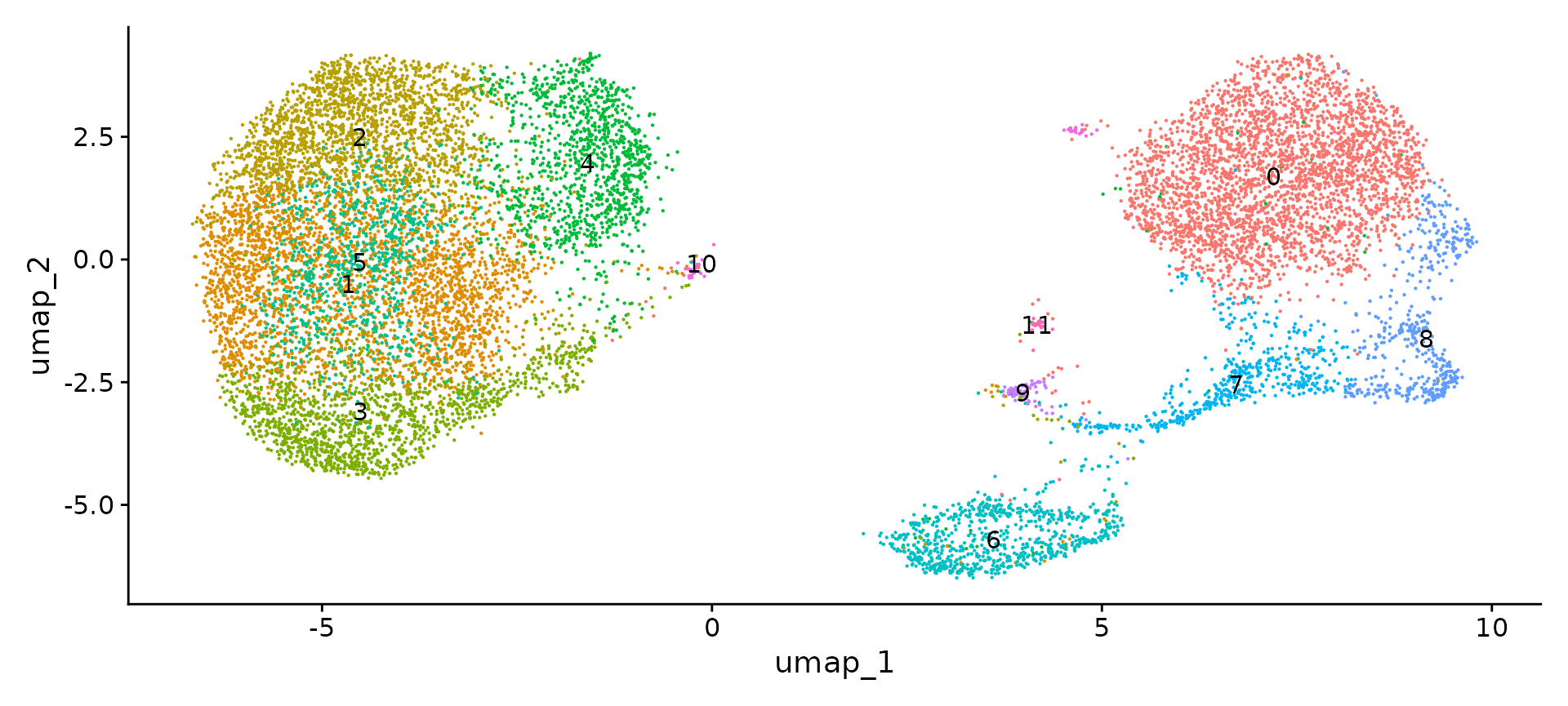
sub type clustering
## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 13572
## Number of edges: 473453
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.8046
## Number of communities: 12
## Elapsed time: 4 seconds
library(ggplot2)
p <- DimPlot(obj, label = T, label.size = 8, reduction = "ref.umap", group.by = "predicted.cluster_full",
alpha = 0.1) + NoLegend()
ggsave(filename = "../output/images/MouseBrain_sketch_clustering.jpg", height = 7, width = 7, plot = p,
quality = 50)
## unnamed.chunk.1 unnamed.chunk.2 unnamed.chunk.3 unnamed.chunk.4
## 1 5.58148 secs 12.51023 secs 98.39153 secs 16.49984 secs
## unnamed.chunk.5 unnamed.chunk.6 unnamed.chunk.7 unnamed.chunk.8
## 1 0.3887007 secs 0.3422155 secs 6.266351 secs 3.129415 secs
## unnamed.chunk.9 unnamed.chunk.10 unnamed.chunk.11 unnamed.chunk.12
## 1 43.44097 secs 2.539532 secs 72.49988 secs 1.903215 secs
## unnamed.chunk.13 unnamed.chunk.14 save.img
## 1 38.27588 secs 0.5088754 secs 1.562802 secs
Session Info
## R version 4.2.0 (2022-04-22)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggplot2_3.4.1 dplyr_1.1.0 biomaRt_2.54.0
## [4] shinyBS_0.61.1 BPCells_0.0.0.9000 Seurat_4.9.9.9035
## [7] SeuratObject_4.9.9.9072 sp_1.6-0
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.3 shinydashboard_0.7.2 spatstat.explore_3.0-6
## [4] reticulate_1.28 tidyselect_1.2.0 RSQLite_2.3.0
## [7] AnnotationDbi_1.60.0 htmlwidgets_1.6.1 grid_4.2.0
## [10] Rtsne_0.16 munsell_0.5.0 codetools_0.2-18
## [13] ragg_1.2.5 ica_1.0-3 DT_0.27
## [16] future_1.32.0 miniUI_0.1.1.1 withr_2.5.0
## [19] spatstat.random_3.1-3 colorspace_2.1-0 progressr_0.13.0
## [22] filelock_1.0.2 Biobase_2.58.0 highr_0.10
## [25] knitr_1.42 stats4_4.2.0 ROCR_1.0-11
## [28] tensor_1.5 listenv_0.9.0 labeling_0.4.2
## [31] GenomeInfoDbData_1.2.9 polyclip_1.10-4 farver_2.1.1
## [34] bit64_4.0.5 rprojroot_2.0.3 parallelly_1.34.0
## [37] vctrs_0.5.2 generics_0.1.3 xfun_0.37
## [40] BiocFileCache_2.6.1 R6_2.5.1 GenomeInfoDb_1.35.15
## [43] hdf5r_1.3.8 bitops_1.0-7 spatstat.utils_3.0-1
## [46] cachem_1.0.7 promises_1.2.0.1 scales_1.2.1
## [49] googlesheets4_1.0.1 gtable_0.3.1 globals_0.16.2
## [52] goftest_1.2-3 spam_2.9-1 rlang_1.0.6
## [55] systemfonts_1.0.4 splines_4.2.0 lazyeval_0.2.2
## [58] gargle_1.3.0 spatstat.geom_3.0-6 yaml_2.3.7
## [61] reshape2_1.4.4 abind_1.4-5 httpuv_1.6.9
## [64] SeuratDisk_0.0.0.9020 tools_4.2.0 Azimuth_0.4.6.9000
## [67] SeuratData_0.2.2.9000 ellipsis_0.3.2 jquerylib_0.1.4
## [70] RColorBrewer_1.1-3 BiocGenerics_0.44.0 ggridges_0.5.4
## [73] Rcpp_1.0.10 plyr_1.8.8 progress_1.2.2
## [76] zlibbioc_1.44.0 purrr_1.0.1 RCurl_1.98-1.10
## [79] prettyunits_1.1.1 deldir_1.0-6 pbapply_1.7-0
## [82] cowplot_1.1.1 S4Vectors_0.36.2 zoo_1.8-11
## [85] ggrepel_0.9.3 cluster_2.1.3 fs_1.6.1
## [88] magrittr_2.0.3 data.table_1.14.8 RSpectra_0.16-1
## [91] scattermore_0.8 lmtest_0.9-40 RANN_2.6.1
## [94] googledrive_2.0.0 fitdistrplus_1.1-8 matrixStats_0.63.0
## [97] hms_1.1.2 patchwork_1.1.2 shinyjs_2.1.0
## [100] mime_0.12 evaluate_0.20 xtable_1.8-4
## [103] XML_3.99-0.13 fastDummies_1.6.3 IRanges_2.32.0
## [106] gridExtra_2.3 compiler_4.2.0 tibble_3.2.0
## [109] KernSmooth_2.23-20 crayon_1.5.2 htmltools_0.5.4
## [112] later_1.3.0 tidyr_1.3.0 DBI_1.1.3
## [115] formatR_1.14 dbplyr_2.3.1 MASS_7.3-56
## [118] rappdirs_0.3.3 Matrix_1.5-3 cli_3.6.0
## [121] parallel_4.2.0 dotCall64_1.0-2 igraph_1.4.1
## [124] GenomicRanges_1.50.2 pkgconfig_2.0.3 pkgdown_2.0.7
## [127] plotly_4.10.1 spatstat.sparse_3.0-0 xml2_1.3.3
## [130] bslib_0.4.2 XVector_0.38.0 stringr_1.5.0
## [133] digest_0.6.31 sctransform_0.3.5 RcppAnnoy_0.0.20
## [136] Biostrings_2.66.0 spatstat.data_3.0-0 rmarkdown_2.20
## [139] cellranger_1.1.0 leiden_0.4.3 uwot_0.1.14
## [142] curl_5.0.0 shiny_1.7.4 lifecycle_1.0.3
## [145] nlme_3.1-157 jsonlite_1.8.4 desc_1.4.2
## [148] viridisLite_0.4.1 fansi_1.0.4 pillar_1.8.1
## [151] lattice_0.20-45 KEGGREST_1.38.0 fastmap_1.1.1
## [154] httr_1.4.5 survival_3.3-1 glue_1.6.2
## [157] png_0.1-8 bit_4.0.5 presto_1.0.0
## [160] stringi_1.7.12 sass_0.4.5 blob_1.2.3
## [163] textshaping_0.3.6 RcppHNSW_0.4.1 memoise_2.0.1
## [166] irlba_2.3.5.1 future.apply_1.10.0






