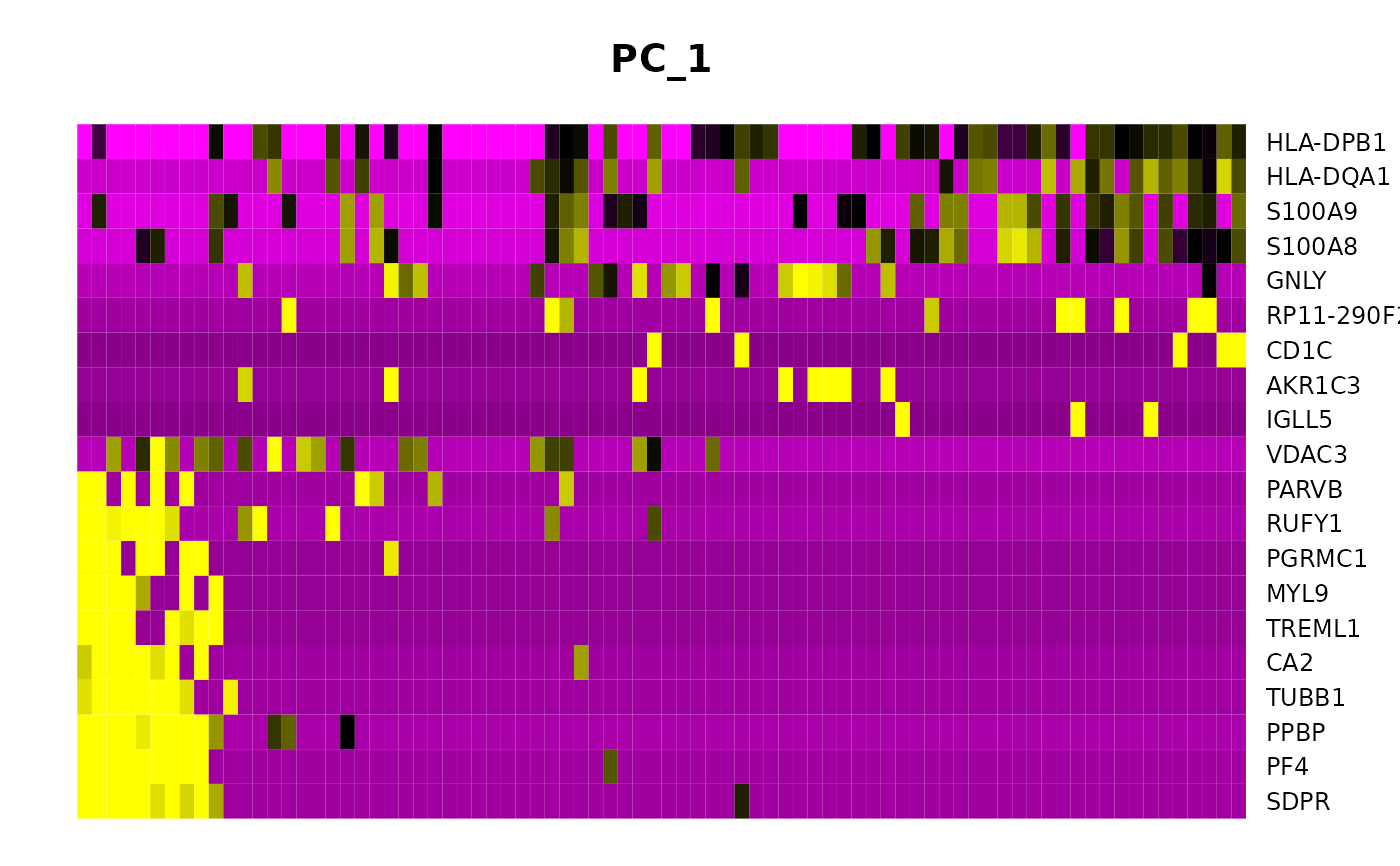
Draws a heatmap focusing on a principal component. Both cells and genes are sorted by their principal component scores. Allows for nice visualization of sources of heterogeneity in the dataset.
DimHeatmap(
object,
dims = 1,
nfeatures = 30,
cells = NULL,
reduction = "pca",
disp.min = -2.5,
disp.max = NULL,
balanced = TRUE,
projected = FALSE,
ncol = NULL,
fast = TRUE,
raster = TRUE,
slot = "scale.data",
assays = NULL,
combine = TRUE
)
PCHeatmap(object, ...)Arguments
- object
Seurat object
- dims
Dimensions to plot
- nfeatures
Number of genes to plot
- cells
A list of cells to plot. If numeric, just plots the top cells.
- reduction
Which dimensional reduction to use
- disp.min
Minimum display value (all values below are clipped)
- disp.max
Maximum display value (all values above are clipped); defaults to 2.5 if
slotis 'scale.data', 6 otherwise- balanced
Plot an equal number of genes with both + and - scores.
- projected
Use the full projected dimensional reduction
- ncol
Number of columns to plot
- fast
If true, use
imageto generate plots; faster than using ggplot2, but not customizable- raster
If true, plot with geom_raster, else use geom_tile. geom_raster may look blurry on some viewing applications such as Preview due to how the raster is interpolated. Set this to FALSE if you are encountering that issue (note that plots may take longer to produce/render).
- slot
Data slot to use, choose from 'raw.data', 'data', or 'scale.data'
- assays
A vector of assays to pull data from
- combine
Combine plots into a single
patchworkedggplot object. IfFALSE, return a list of ggplot objects- ...
Extra parameters passed to
DimHeatmap
Value
No return value by default. If using fast = FALSE, will return a
patchworked ggplot object if combine = TRUE, otherwise
returns a list of ggplot objects
See also
Examples
data("pbmc_small")
DimHeatmap(object = pbmc_small)
#> Warning: Requested number is larger than the number of available items (20). Setting to 20.