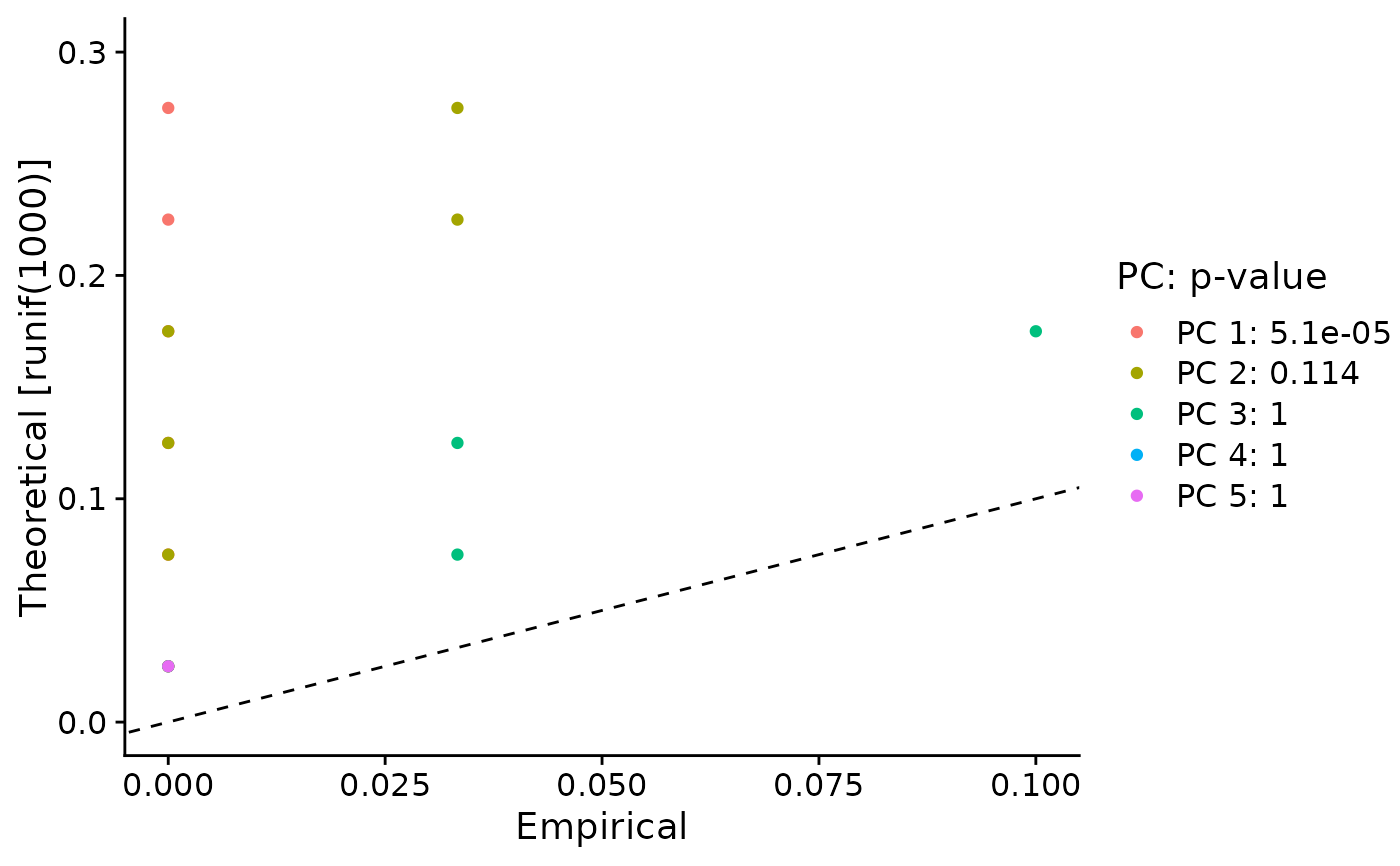
Plots the results of the JackStraw analysis for PCA significance. For each PC, plots a QQ-plot comparing the distribution of p-values for all genes across each PC, compared with a uniform distribution. Also determines a p-value for the overall significance of each PC (see Details).
JackStrawPlot(
object,
dims = 1:5,
cols = NULL,
reduction = "pca",
xmax = 0.1,
ymax = 0.3
)Arguments
- object
Seurat object
- dims
Dims to plot
- cols
Vector of colors, each color corresponds to an individual PC. This may also be a single character or numeric value corresponding to a palette as specified by
brewer.pal.info. By default, ggplot2 assigns colors. We also include a number of palettes from the pals package. SeeDiscretePalettefor details.- reduction
reduction to pull jackstraw info from
- xmax
X-axis maximum on each QQ plot.
- ymax
Y-axis maximum on each QQ plot.
Value
A ggplot object
Details
Significant PCs should show a p-value distribution (black curve) that is strongly skewed to the left compared to the null distribution (dashed line) The p-value for each PC is based on a proportion test comparing the number of genes with a p-value below a particular threshold (score.thresh), compared with the proportion of genes expected under a uniform distribution of p-values.
See also
Examples
data("pbmc_small")
JackStrawPlot(object = pbmc_small)
#> Warning: Removed 83 rows containing missing values or values outside the scale range (`geom_point()`).