Demultiplexing with hashtag oligos (HTOs)
Compiled: 2023-03-27
Source:vignettes/hashing_vignette.Rmdhashing_vignette.RmdDeveloped in collaboration with the Technology Innovation Group at NYGC, Cell Hashing uses oligo-tagged antibodies against ubiquitously expressed surface proteins to place a “sample barcode” on each single cell, enabling different samples to be multiplexed together and run in a single experiment. For more information, please refer to this paper.
This vignette will give a brief demonstration on how to work with data produced with Cell Hashing in Seurat. Applied to two datasets, we can successfully demultiplex cells to their the original sample-of-origin, and identify cross-sample doublets.
HTODemux() implements the following procedure:- We perform a k-medoid clustering on the normalized HTO values, which initially separates cells into K(# of samples)+1 clusters.
- We calculate a ‘negative’ distribution for HTO. For each HTO, we use the cluster with the lowest average value as the negative group.
- For each HTO, we fit a negative binomial distribution to the negative cluster. We use the 0.99 quantile of this distribution as a threshold.
- Based on these thresholds, each cell is classified as positive or negative for each HTO.
- Cells that are positive for more than one HTOs are annotated as doublets.
8-HTO dataset from human PBMCs
- Data represent peripheral blood mononuclear cells (PBMCs) from eight different donors.
- Cells from each donor are uniquely labeled, using CD45 as a hashing antibody.
- Samples were subsequently pooled, and run on a single lane of the the 10X Chromium v2 system.
- You can download the count matrices for RNA and HTO here, or the FASTQ files from GEO
Basic setup
Load packages
Read in data
# Load in the UMI matrix
pbmc.umis <- readRDS("../data/pbmc_umi_mtx.rds")
# For generating a hashtag count matrix from FASTQ files, please refer to
# https://github.com/Hoohm/CITE-seq-Count. Load in the HTO count matrix
pbmc.htos <- readRDS("../data/pbmc_hto_mtx.rds")
# Select cell barcodes detected by both RNA and HTO In the example datasets we have already
# filtered the cells for you, but perform this step for clarity.
joint.bcs <- intersect(colnames(pbmc.umis), colnames(pbmc.htos))
# Subset RNA and HTO counts by joint cell barcodes
pbmc.umis <- pbmc.umis[, joint.bcs]
pbmc.htos <- as.matrix(pbmc.htos[, joint.bcs])
# Confirm that the HTO have the correct names
rownames(pbmc.htos)## [1] "HTO_A" "HTO_B" "HTO_C" "HTO_D" "HTO_E" "HTO_F" "HTO_G" "HTO_H"Setup Seurat object and add in the HTO data
# Setup Seurat object
pbmc.hashtag <- CreateSeuratObject(counts = pbmc.umis)
# Normalize RNA data with log normalization
pbmc.hashtag <- NormalizeData(pbmc.hashtag)
# Find and scale variable features
pbmc.hashtag <- FindVariableFeatures(pbmc.hashtag, selection.method = "mean.var.plot")
pbmc.hashtag <- ScaleData(pbmc.hashtag, features = VariableFeatures(pbmc.hashtag))Adding HTO data as an independent assay
You can read more about working with multi-modal data here
# Add HTO data as a new assay independent from RNA
pbmc.hashtag[["HTO"]] <- CreateAssayObject(counts = pbmc.htos)
# Normalize HTO data, here we use centered log-ratio (CLR) transformation
pbmc.hashtag <- NormalizeData(pbmc.hashtag, assay = "HTO", normalization.method = "CLR")Demultiplex cells based on HTO enrichment
Here we use the Seurat function HTODemux() to assign single cells back to their sample origins.
# If you have a very large dataset we suggest using k_function = 'clara'. This is a k-medoid
# clustering function for large applications You can also play with additional parameters (see
# documentation for HTODemux()) to adjust the threshold for classification Here we are using
# the default settings
pbmc.hashtag <- HTODemux(pbmc.hashtag, assay = "HTO", positive.quantile = 0.99)Visualize demultiplexing results
Output from running HTODemux() is saved in the object metadata. We can visualize how many cells are classified as singlets, doublets and negative/ambiguous cells.
# Global classification results
table(pbmc.hashtag$HTO_classification.global)##
## Doublet Negative Singlet
## 2598 346 13972Visualize enrichment for selected HTOs with ridge plots
# Group cells based on the max HTO signal
Idents(pbmc.hashtag) <- "HTO_maxID"
RidgePlot(pbmc.hashtag, assay = "HTO", features = rownames(pbmc.hashtag[["HTO"]])[1:2], ncol = 2)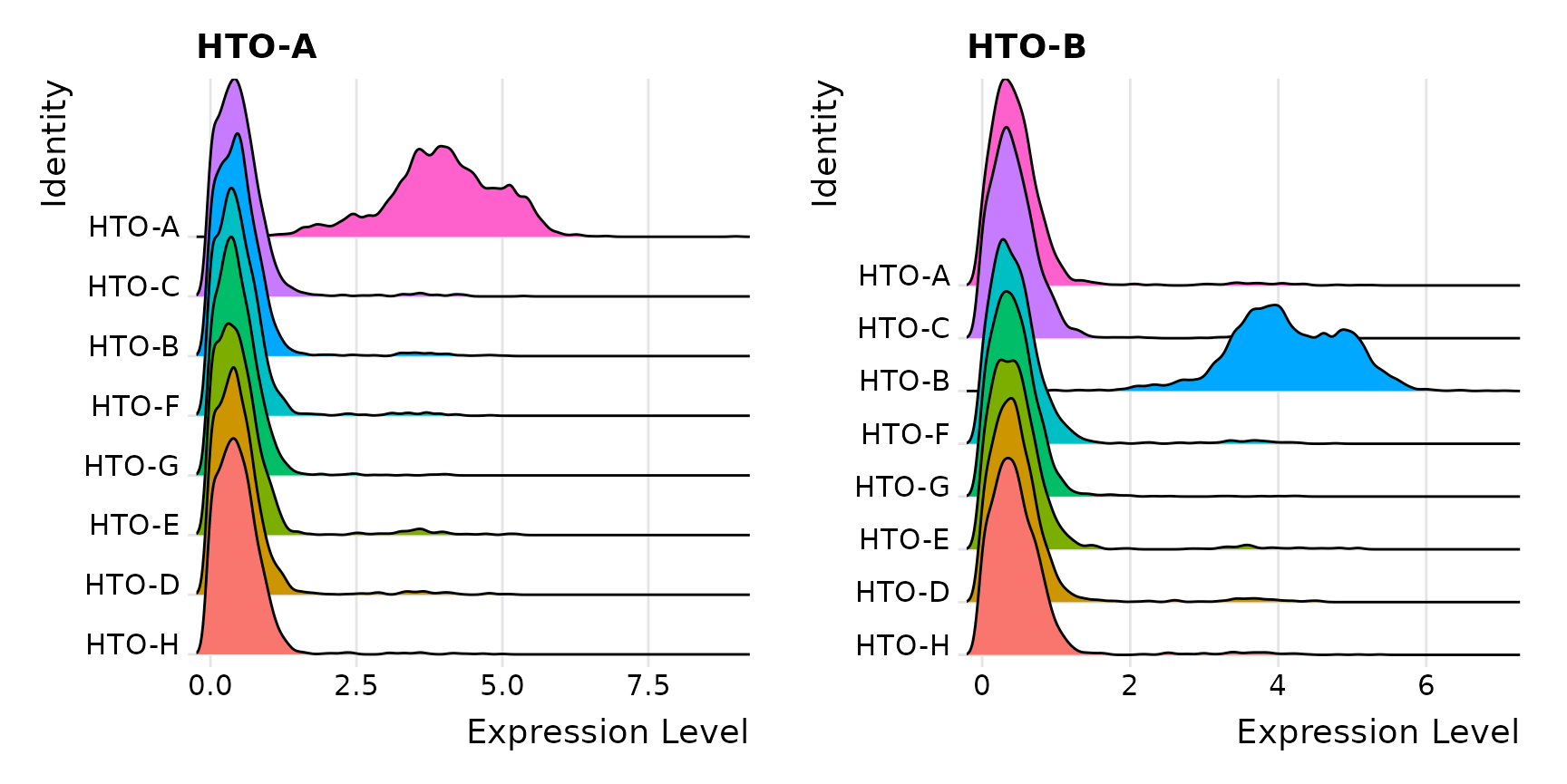
Visualize pairs of HTO signals to confirm mutual exclusivity in singlets
FeatureScatter(pbmc.hashtag, feature1 = "hto_HTO-A", feature2 = "hto_HTO-B")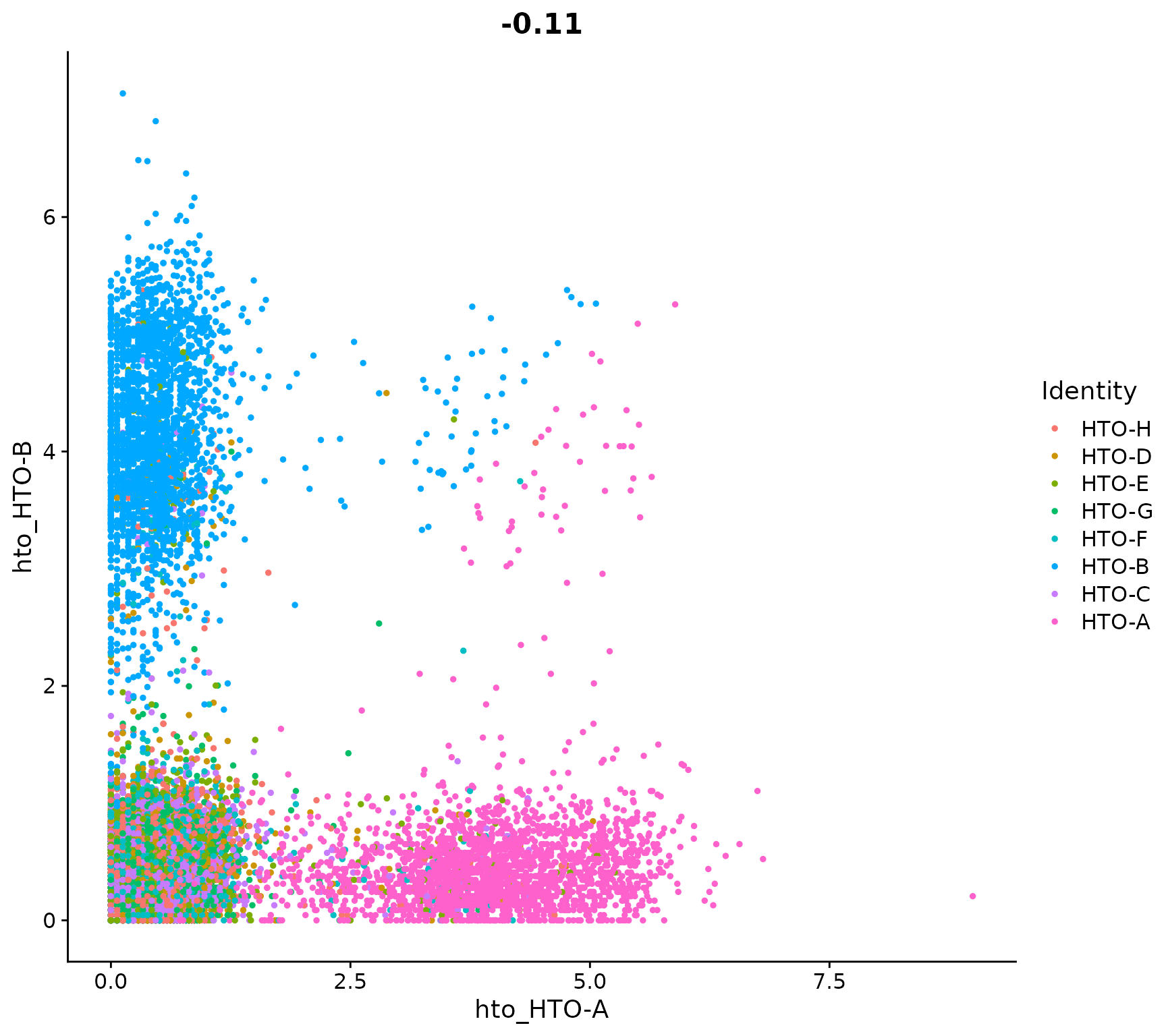
Compare number of UMIs for singlets, doublets and negative cells
Idents(pbmc.hashtag) <- "HTO_classification.global"
VlnPlot(pbmc.hashtag, features = "nCount_RNA", pt.size = 0.1, log = TRUE)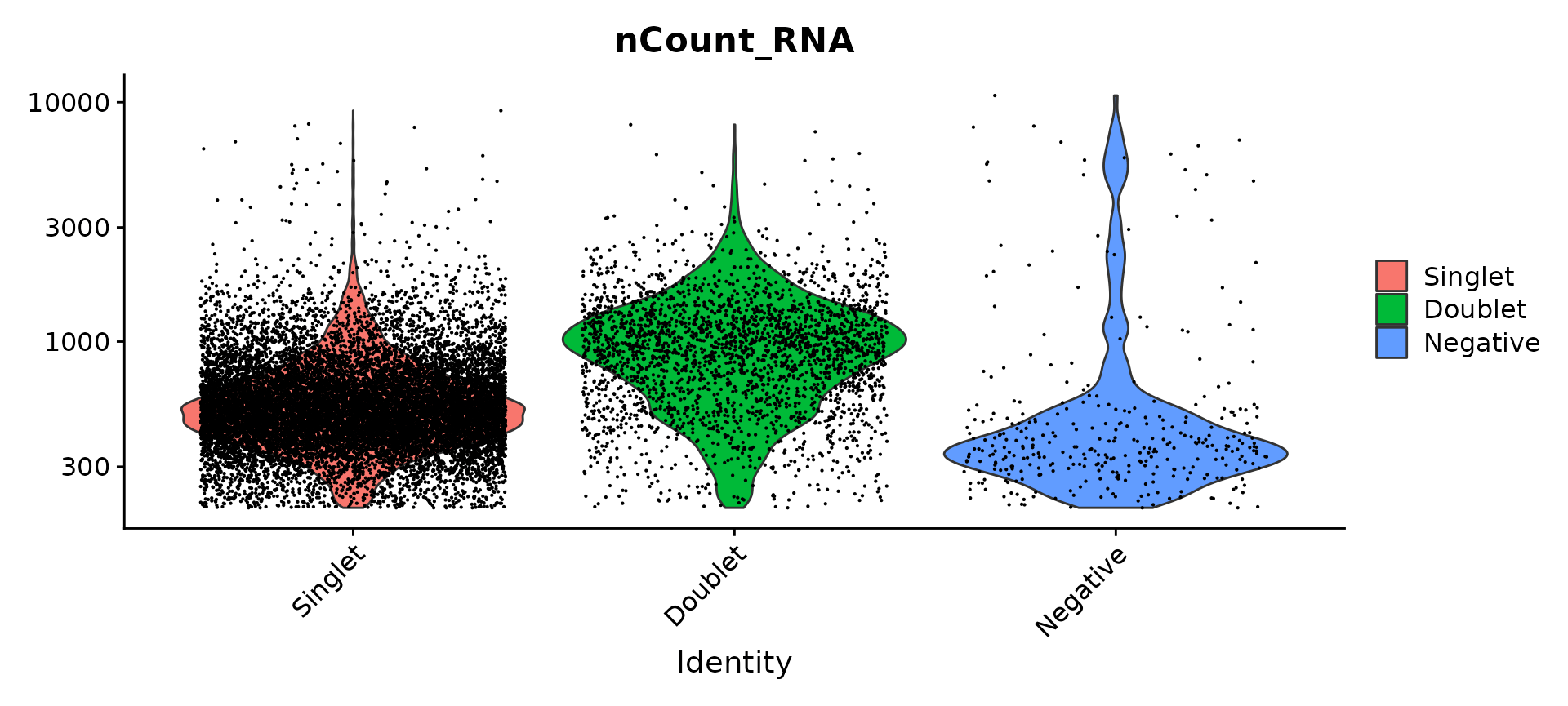
Generate a two dimensional tSNE embedding for HTOs.Here we are grouping cells by singlets and doublets for simplicity.
# First, we will remove negative cells from the object
pbmc.hashtag.subset <- subset(pbmc.hashtag, idents = "Negative", invert = TRUE)
# Calculate a tSNE embedding of the HTO data
DefaultAssay(pbmc.hashtag.subset) <- "HTO"
pbmc.hashtag.subset <- ScaleData(pbmc.hashtag.subset, features = rownames(pbmc.hashtag.subset),
verbose = FALSE)
pbmc.hashtag.subset <- RunPCA(pbmc.hashtag.subset, features = rownames(pbmc.hashtag.subset), approx = FALSE)
pbmc.hashtag.subset <- RunTSNE(pbmc.hashtag.subset, dims = 1:8, perplexity = 100)
DimPlot(pbmc.hashtag.subset)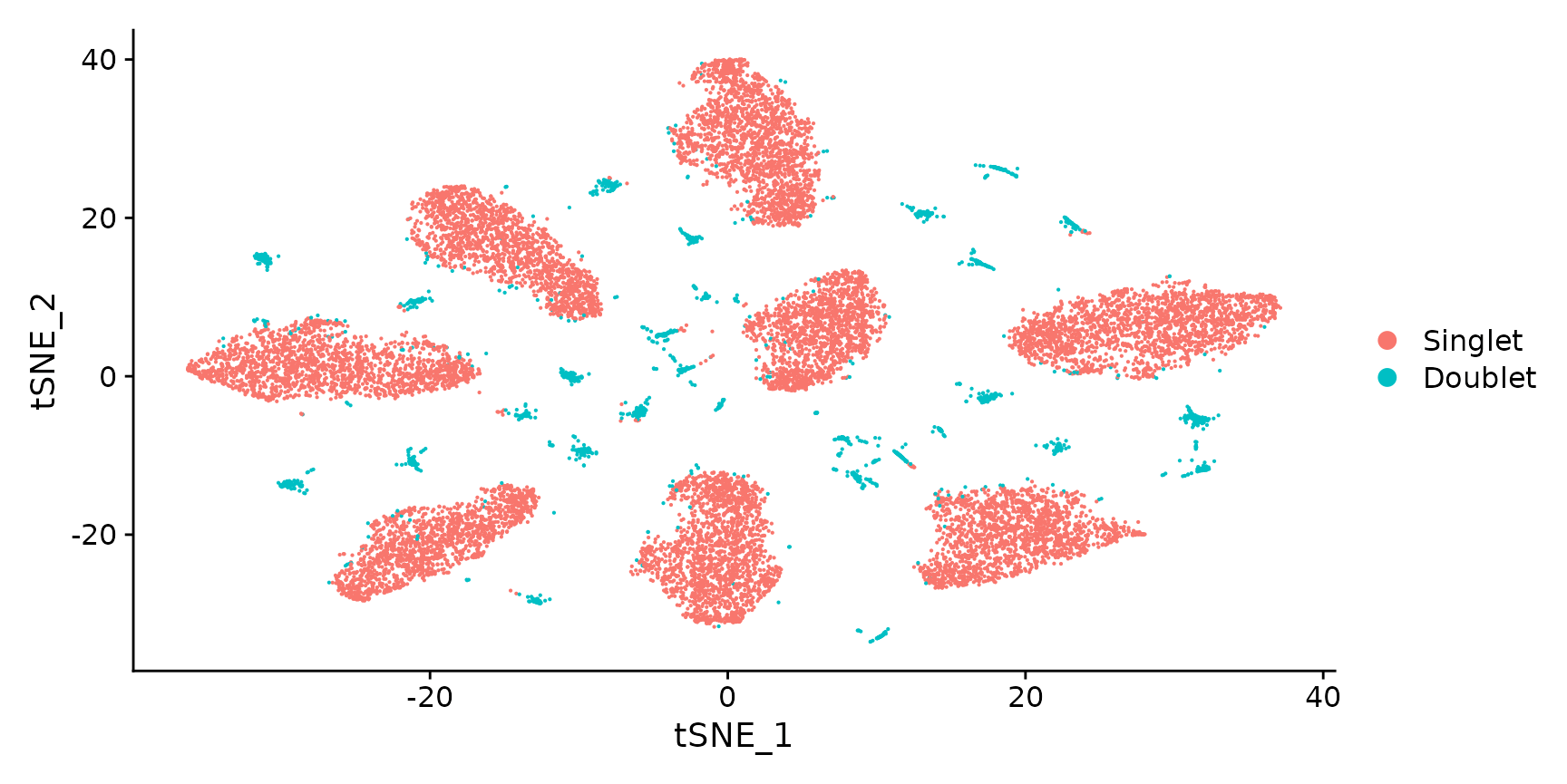
# You can also visualize the more detailed classification result by running Idents(object) <-
# 'HTO_classification' before plotting. Here, you can see that each of the small clouds on the
# tSNE plot corresponds to one of the 28 possible doublet combinations.Create an HTO heatmap, based on Figure 1C in the Cell Hashing paper.
# To increase the efficiency of plotting, you can subsample cells using the num.cells argument
HTOHeatmap(pbmc.hashtag, assay = "HTO", ncells = 5000)
Cluster and visualize cells using the usual scRNA-seq workflow, and examine for the potential presence of batch effects.
# Extract the singlets
pbmc.singlet <- subset(pbmc.hashtag, idents = "Singlet")
# Select the top 1000 most variable features
pbmc.singlet <- FindVariableFeatures(pbmc.singlet, selection.method = "mean.var.plot")
# Scaling RNA data, we only scale the variable features here for efficiency
pbmc.singlet <- ScaleData(pbmc.singlet, features = VariableFeatures(pbmc.singlet))
# Run PCA
pbmc.singlet <- RunPCA(pbmc.singlet, features = VariableFeatures(pbmc.singlet))
# We select the top 10 PCs for clustering and tSNE based on PCElbowPlot
pbmc.singlet <- FindNeighbors(pbmc.singlet, reduction = "pca", dims = 1:10)
pbmc.singlet <- FindClusters(pbmc.singlet, resolution = 0.6, verbose = FALSE)
pbmc.singlet <- RunTSNE(pbmc.singlet, reduction = "pca", dims = 1:10)
# Projecting singlet identities on TSNE visualization
DimPlot(pbmc.singlet, group.by = "HTO_classification")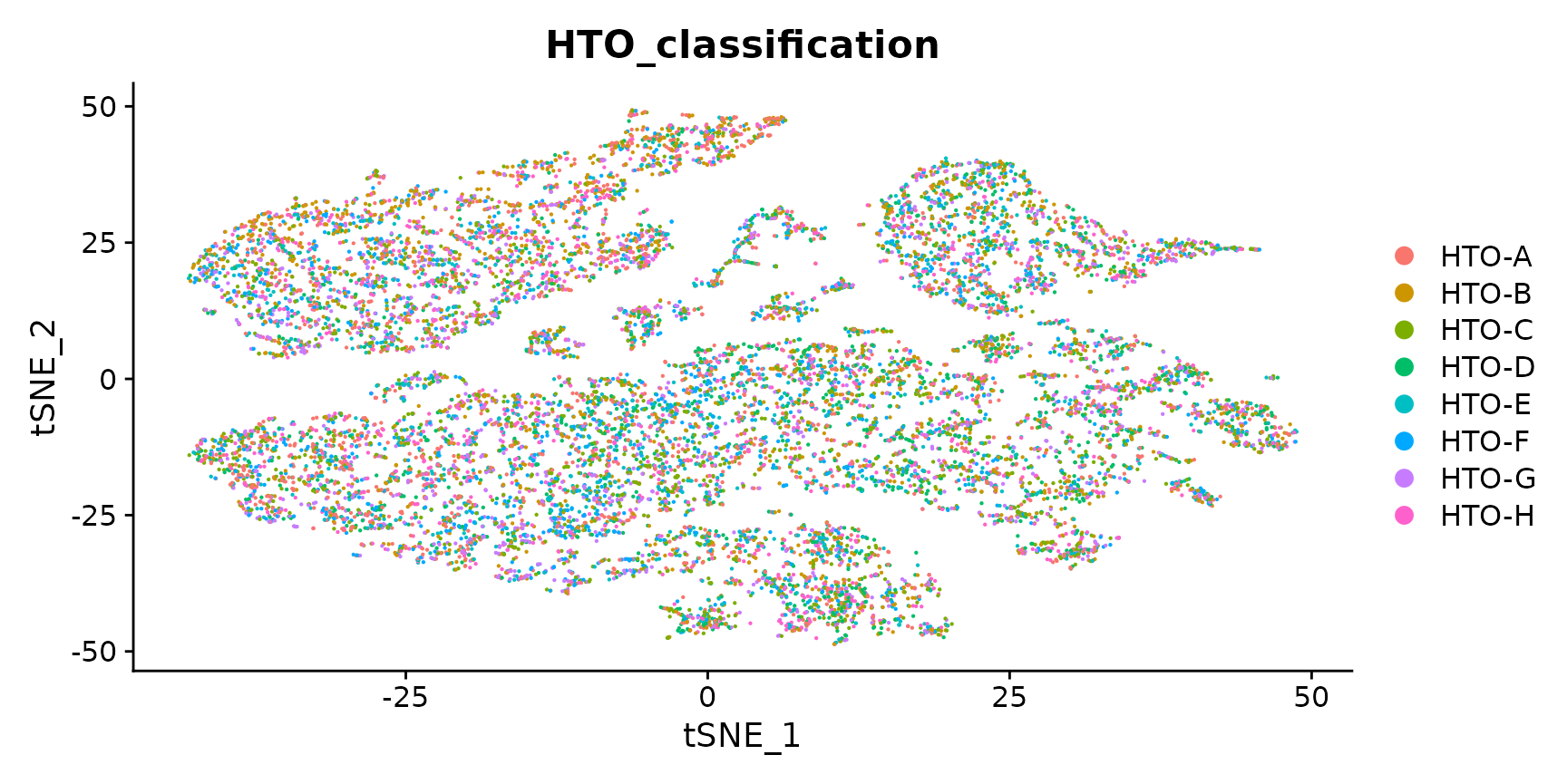
12-HTO dataset from four human cell lines
- Data represent single cells collected from four cell lines: HEK, K562, KG1 and THP1
- Each cell line was further split into three samples (12 samples in total).
- Each sample was labeled with a hashing antibody mixture (CD29 and CD45), pooled, and run on a single lane of 10X.
- Based on this design, we should be able to detect doublets both across and within cell types
- You can download the count matrices for RNA and HTO here, and are available on GEO here
Create Seurat object, add HTO data and perform normalization
# Read in UMI count matrix for RNA
hto12.umis <- readRDS("../data/hto12_umi_mtx.rds")
# Read in HTO count matrix
hto12.htos <- readRDS("../data/hto12_hto_mtx.rds")
# Select cell barcodes detected in both RNA and HTO
cells.use <- intersect(rownames(hto12.htos), colnames(hto12.umis))
# Create Seurat object and add HTO data
hto12 <- CreateSeuratObject(counts = hto12.umis[, cells.use], min.features = 300)
hto12[["HTO"]] <- CreateAssayObject(counts = t(x = hto12.htos[colnames(hto12), 1:12]))
# Normalize data
hto12 <- NormalizeData(hto12)
hto12 <- NormalizeData(hto12, assay = "HTO", normalization.method = "CLR")Demultiplex data
hto12 <- HTODemux(hto12, assay = "HTO", positive.quantile = 0.99)Visualize demultiplexing results
Distribution of selected HTOs grouped by classification, displayed by ridge plots
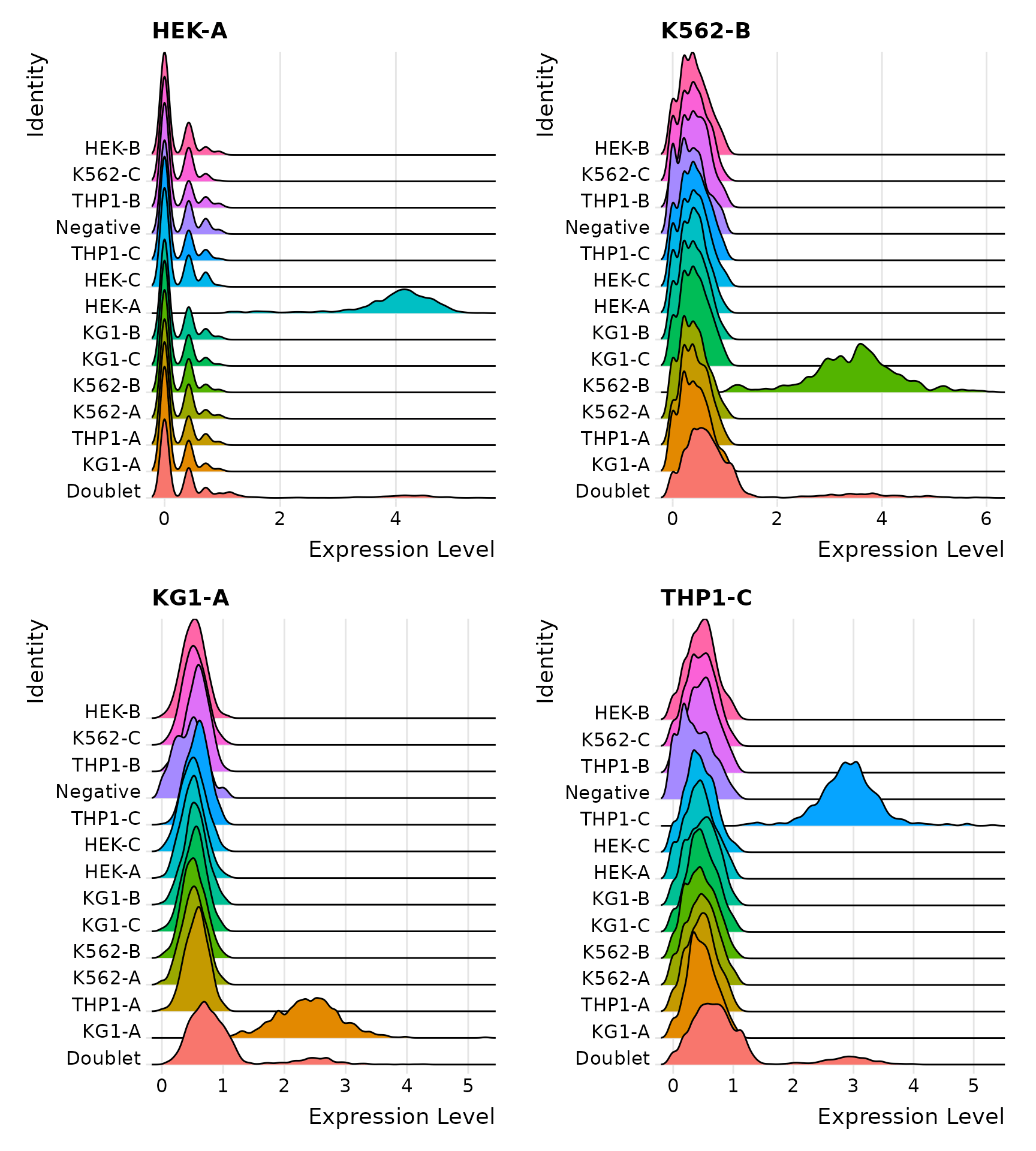
Visualize HTO signals in a heatmap
HTOHeatmap(hto12, assay = "HTO")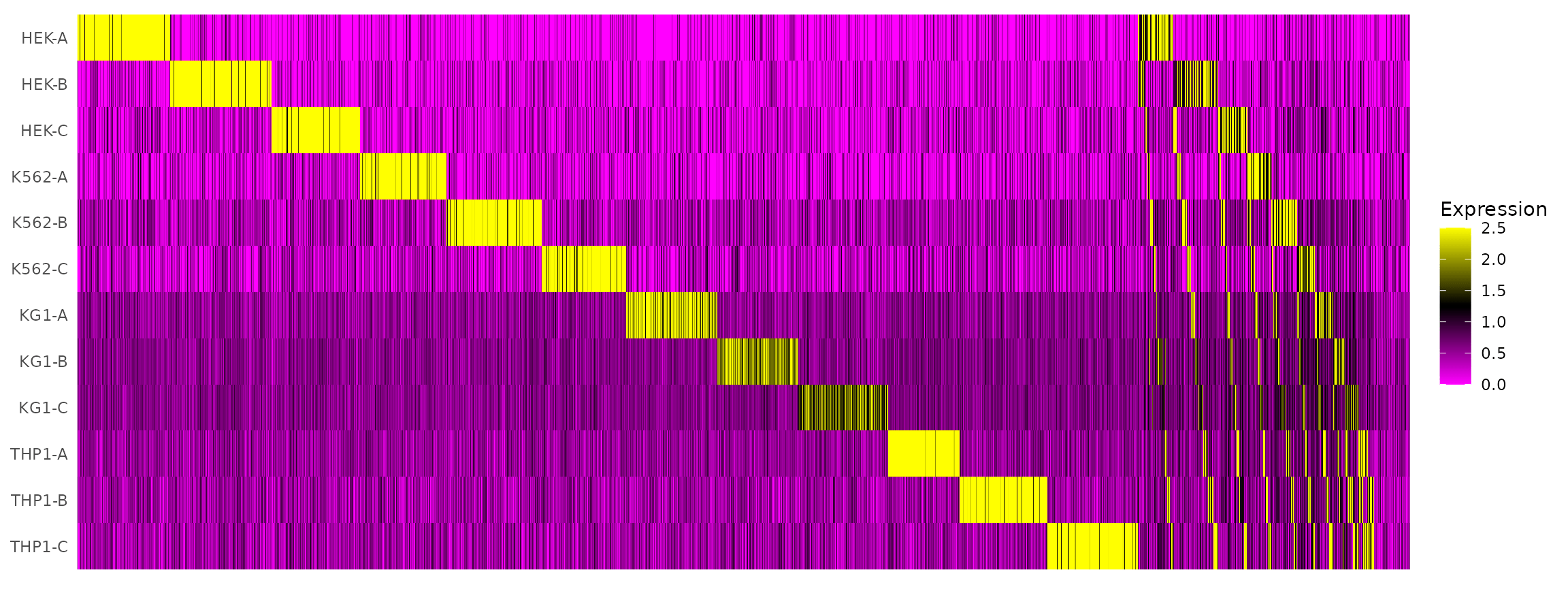
Visualize RNA clustering
# Remove the negative cells
hto12 <- subset(hto12, idents = "Negative", invert = TRUE)
# Run PCA on most variable features
hto12 <- FindVariableFeatures(hto12, selection.method = "mean.var.plot")
hto12 <- ScaleData(hto12, features = VariableFeatures(hto12))
hto12 <- RunPCA(hto12)
hto12 <- RunTSNE(hto12, dims = 1:5, perplexity = 100)
DimPlot(hto12) + NoLegend()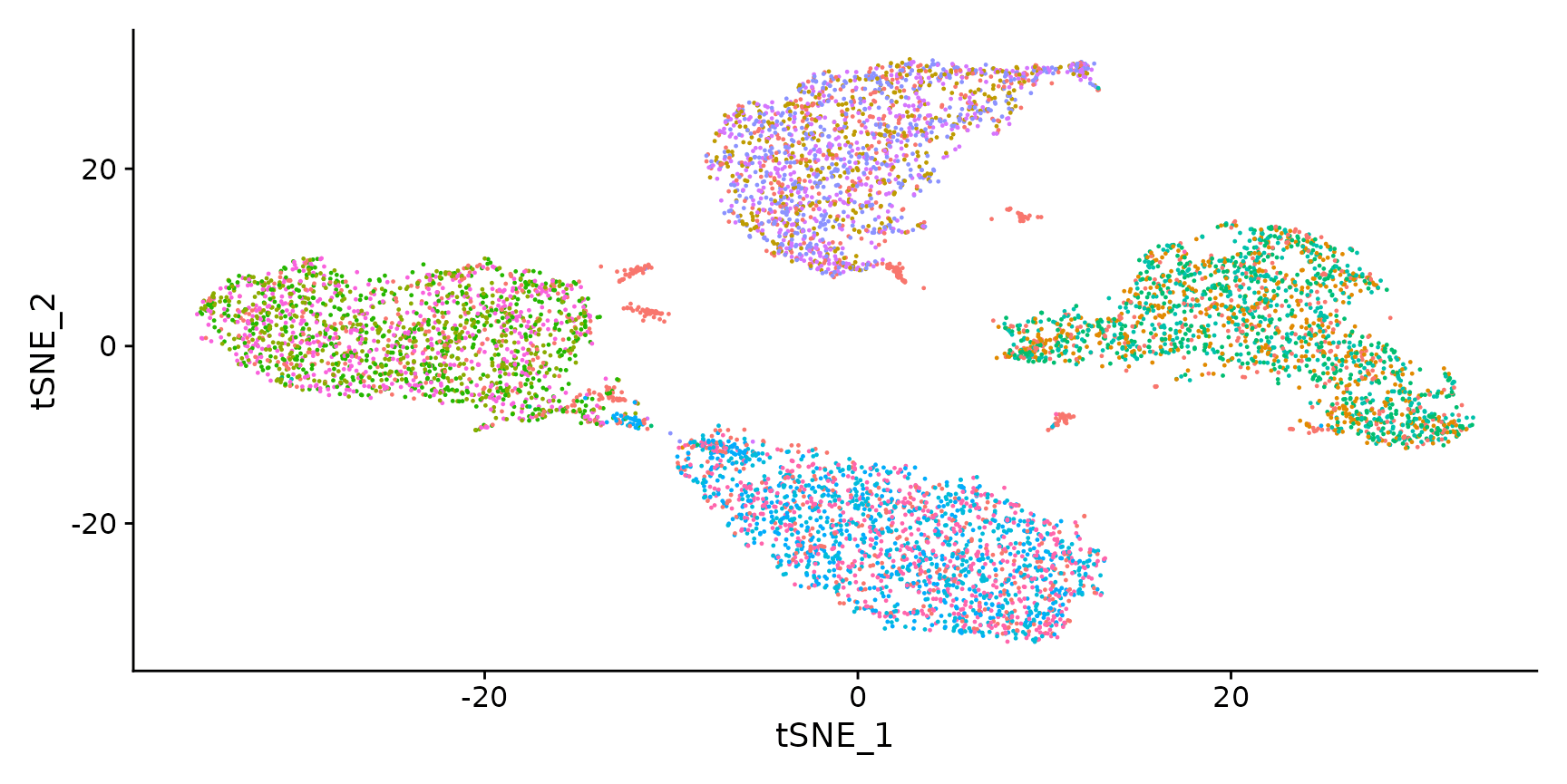
Session Info
## R version 4.2.0 (2022-04-22)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] SeuratObject_4.1.3 Seurat_4.3.0
##
## loaded via a namespace (and not attached):
## [1] Rtsne_0.16 colorspace_2.1-0 deldir_1.0-6
## [4] ellipsis_0.3.2 ggridges_0.5.4 rprojroot_2.0.3
## [7] fs_1.6.1 spatstat.data_3.0-0 farver_2.1.1
## [10] leiden_0.4.3 listenv_0.9.0 ggrepel_0.9.3
## [13] fansi_1.0.4 codetools_0.2-18 splines_4.2.0
## [16] cachem_1.0.7 knitr_1.42 polyclip_1.10-4
## [19] jsonlite_1.8.4 ica_1.0-3 cluster_2.1.3
## [22] png_0.1-8 uwot_0.1.14 spatstat.sparse_3.0-0
## [25] shiny_1.7.4 sctransform_0.3.5 compiler_4.2.0
## [28] httr_1.4.5 Matrix_1.5-3 fastmap_1.1.1
## [31] lazyeval_0.2.2 cli_3.6.0 later_1.3.0
## [34] formatR_1.14 htmltools_0.5.4 tools_4.2.0
## [37] igraph_1.4.1 gtable_0.3.1 glue_1.6.2
## [40] RANN_2.6.1 reshape2_1.4.4 dplyr_1.1.0
## [43] Rcpp_1.0.10 scattermore_0.8 jquerylib_0.1.4
## [46] pkgdown_2.0.7 vctrs_0.5.2 nlme_3.1-157
## [49] spatstat.explore_3.0-6 progressr_0.13.0 lmtest_0.9-40
## [52] spatstat.random_3.1-3 xfun_0.37 stringr_1.5.0
## [55] globals_0.16.2 mime_0.12 miniUI_0.1.1.1
## [58] lifecycle_1.0.3 irlba_2.3.5.1 goftest_1.2-3
## [61] future_1.31.0 MASS_7.3-56 zoo_1.8-11
## [64] scales_1.2.1 ragg_1.2.5 promises_1.2.0.1
## [67] spatstat.utils_3.0-1 parallel_4.2.0 RColorBrewer_1.1-3
## [70] yaml_2.3.7 memoise_2.0.1 reticulate_1.28
## [73] pbapply_1.7-0 gridExtra_2.3 ggplot2_3.4.1
## [76] sass_0.4.5 stringi_1.7.12 highr_0.10
## [79] desc_1.4.2 rlang_1.0.6 pkgconfig_2.0.3
## [82] systemfonts_1.0.4 matrixStats_0.63.0 evaluate_0.20
## [85] lattice_0.20-45 tensor_1.5 ROCR_1.0-11
## [88] purrr_1.0.1 labeling_0.4.2 patchwork_1.1.2
## [91] htmlwidgets_1.6.1 cowplot_1.1.1 tidyselect_1.2.0
## [94] parallelly_1.34.0 RcppAnnoy_0.0.20 plyr_1.8.8
## [97] magrittr_2.0.3 R6_2.5.1 generics_0.1.3
## [100] withr_2.5.0 pillar_1.8.1 fitdistrplus_1.1-8
## [103] abind_1.4-5 survival_3.3-1 sp_1.6-0
## [106] tibble_3.1.8 future.apply_1.10.0 KernSmooth_2.23-20
## [109] utf8_1.2.3 spatstat.geom_3.0-6 plotly_4.10.1
## [112] rmarkdown_2.20 grid_4.2.0 data.table_1.14.8
## [115] digest_0.6.31 xtable_1.8-4 tidyr_1.3.0
## [118] httpuv_1.6.9 textshaping_0.3.6 munsell_0.5.0
## [121] viridisLite_0.4.1 bslib_0.4.2