2025
Jiang L*, Dalgarno C*, Papalexi E, Mascio I, Wessels HH, Yun H, Iremadze N, Lithwick-Yanai G, Lipson D, Satija R
Nature Cell Biology. 2025
Watch Longda Jiang and Carol Dalgarno's CEGS Virtual Series talk on this work
Blair J, Hartman A, Zenk F, Wahle P, Brancati G, Dalgarno C, Treutlein B, Satija R
Nature Communications. 2025
Watch John Blair's CEGS Virtual Series talk on this work
2024
Kowalski M*, Wessels H*, Linder J*, Dalgarno C, Mascio I, Choudhary S, Hartman A, Hao Y, Kundaje A, Satija R
Cell. 2024
Watch Madeline Kowalski and Harm Wessels's CEGS Virtual Series talk on this work
Lotfollahi M*, Hao Y*, Theis FJ#, Satija R#
Cell. 2024

Hartman A, Satija R
eLife. 2024

Jiang L*, Dalgarno C*, Papalexi E, Mascio I, Wessels HH, Yun H, Iremadze N, Lithwick-Yanai G, Lipson D, Satija R
bioRxiv. 2024

2023
Zhang B*, Upadhyay R*, Hao Y, Samanovic M, Herati R, Blair J, Axelrad J, Mulligan M, Littman D#, Satija R#
Nature Immunology. 2023
Watch Bingjie Zhang's CEGS Virtual Series talk on this work
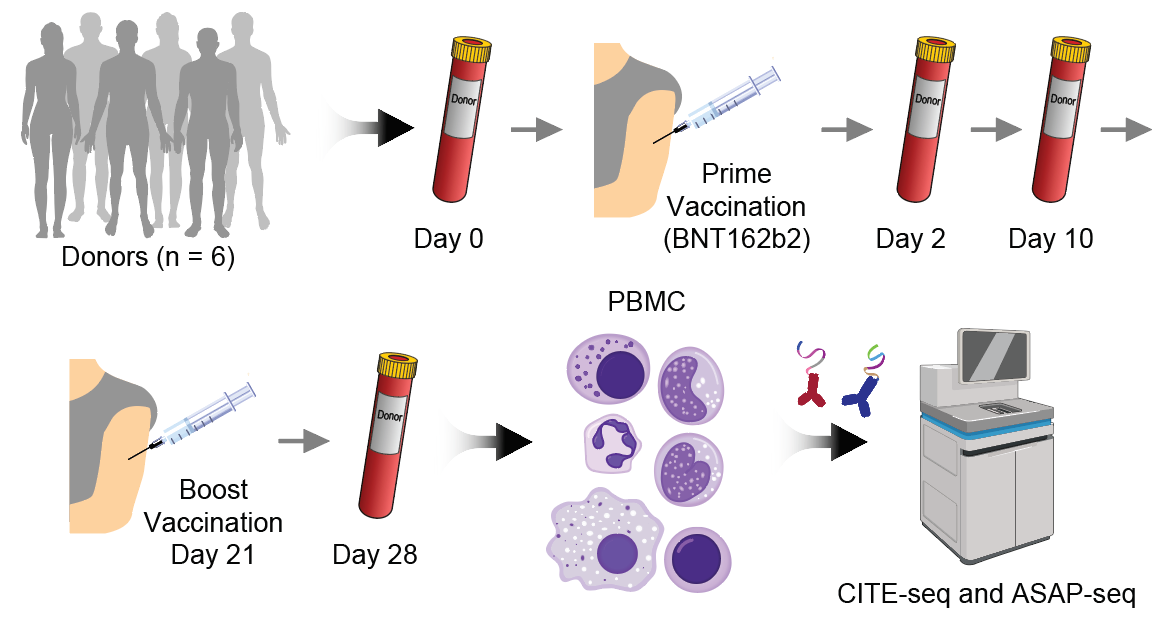
Hao Y, Stuart T, Kowalski M, Choudhary S, Hoffman P, Hartman A, Srivastava A, Molla G, Madad S, Fernandez-Granda C, Satija R
Nature Biotechnology. 2023
Watch Yuhan Hao's CEGS Virtual Series talk on this work

2022
Wessels H*, Mendez-Mancilla A*, Hao Y, Papalexi E, Mauck W, Lu L, Morris J, Mimitou E, Smibert P, Sanjana N#, Satija R#
Nature Methods. 2022
Watch Harm Wessels's CEGS Virtual Series talk on this work
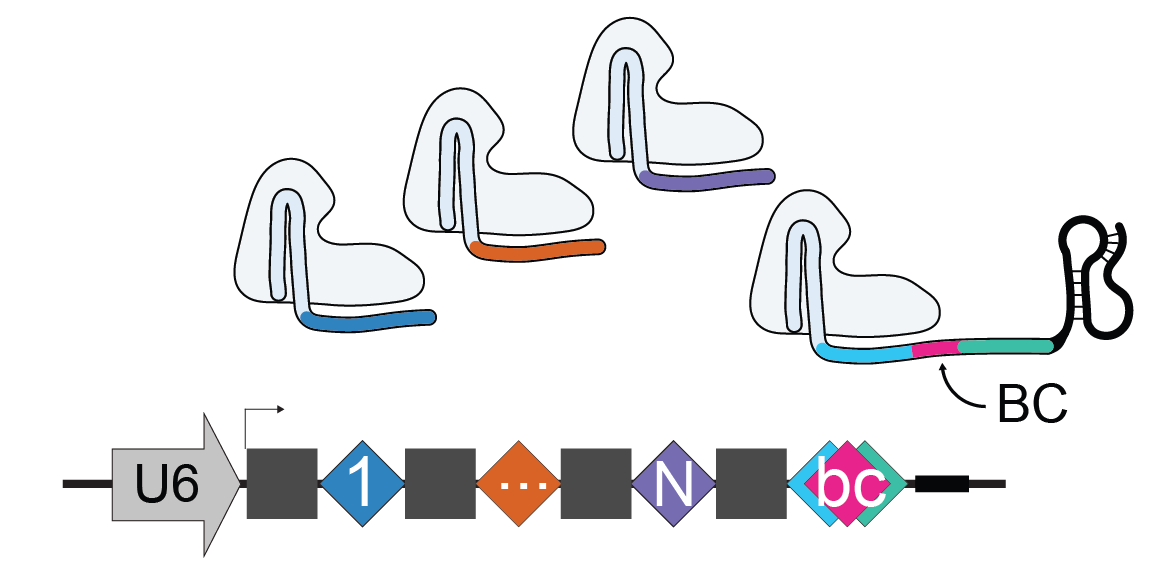
Stuart T, Hao S, Zhang B, Mekerishvili L, Landau D, Maniatis S, Satija R, Raimondi I
Nature Biotechnology. 2022
Watch Tim Stuart's CEGS Virtual Series talk on this work
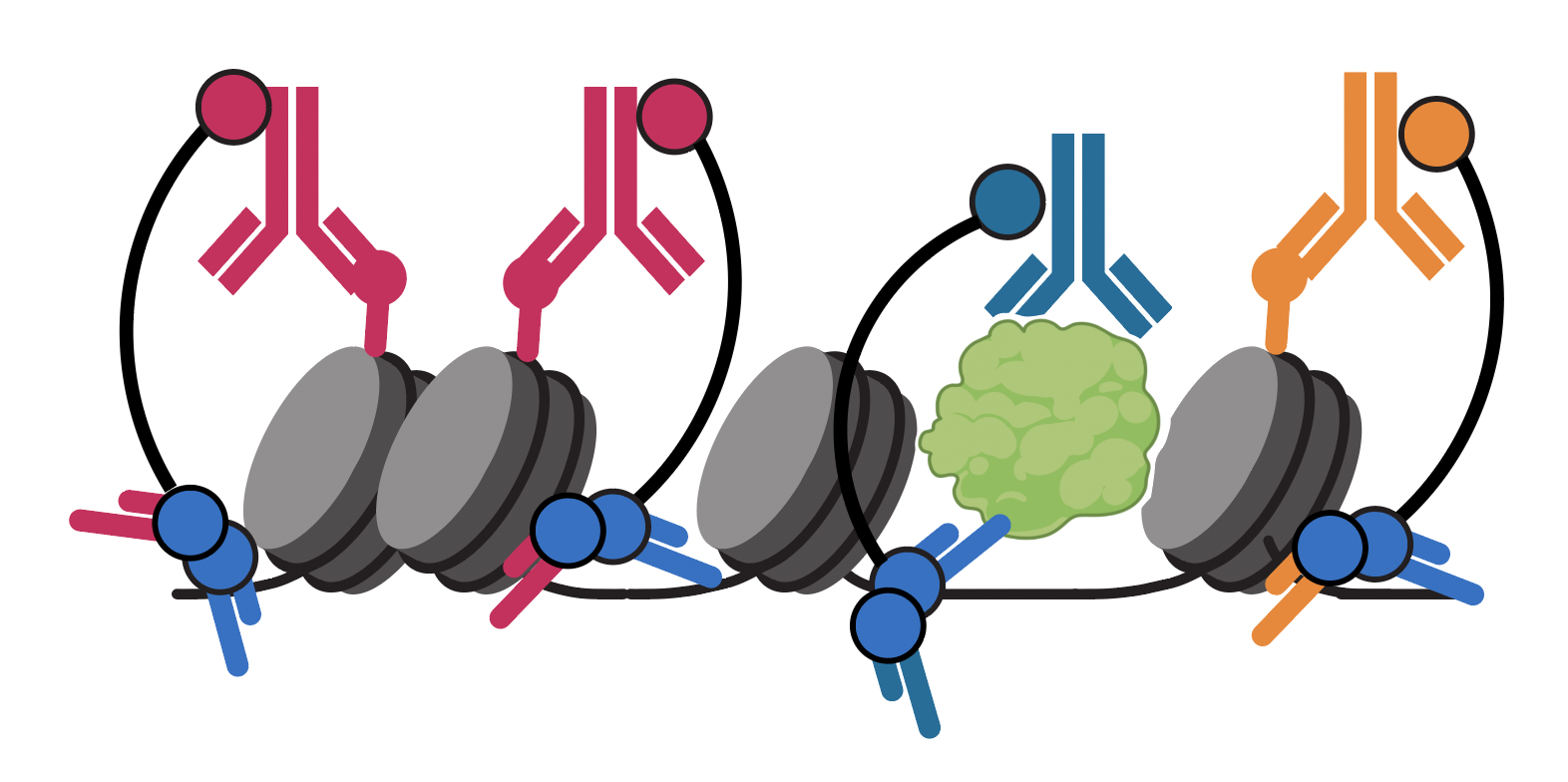
Zhang B*, Srivastava A*, Mimitou E, Stuart T, Raimondi I, Hao Y, Smibert P, Satija R
Nature Biotechnology. 2022
Watch Bingjie Zhang and Avi Srivastava's CEGS Virtual Series talk on this work
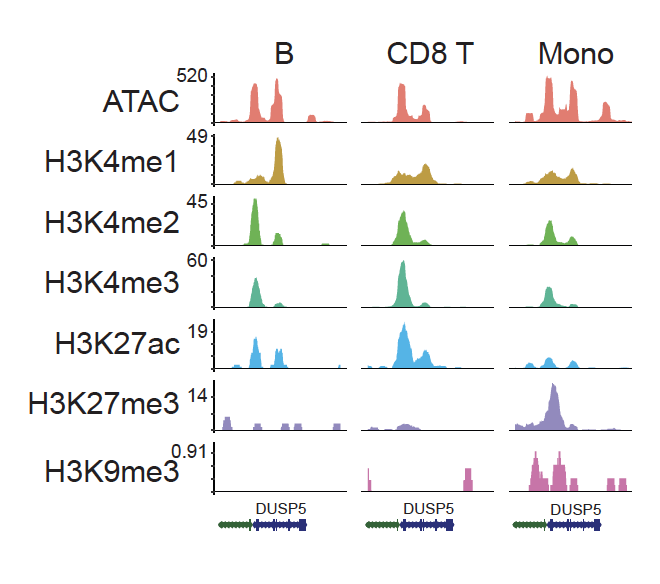
Choudhary S, Satija R
Genome Biology. 2022
2021
Stuart T, Srivastava A, Madad S, Lareau C, Satija R
Nature Methods. 2021

Missarova A, Jain J, Butler A, Ghazanfar S, Stuart T, Brusko M, Wasserfall C, Nick H, Brusko T, Atkinson M, Satija R#, Marioni J#
bioRxiv. 2021
Mimitou E, Lareau C, Chen K, Zorzetto-Fernandes A, Hao Y, Takeshima Y, Luo W, Huang T, Yeung B, Papalexi E, Thakore P, Kibayashi T, Wing J, Hata M, Satija R, Nazor K, Sakaguchi S, Ludwig L, Sankaran V, Regev A, Smibert P
Nature Biotechnology. 2021
Hao Y*, Hao S*, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zagar M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LB, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P*, Satija R*
Cell. 2021
Papalexi E, Mimitou E, Butler A, Foster S, Bracken B, Mauck WM, Wessels H, Yeung BZ, Smibert P, Satija R
Nature Genetics. 2021
Kaufmann KB, Zeng AGX, Coyaud E, Garcia-Prat L, Papalexi E, Murison A, Laurent EMN, Chan-Seng-Yue M, Gan OI, Pan K, McLeod J, Boutzen H, Zandi S, Takayanagi S, Satija R, Raught B, Xie SZ, Dick J
Nature Immunology. 2021
2020
Wu L, Hollinshead KER, Hao Y, Au C, Kroehling L, Ng, C, Woan-Yu L, Li D, Silva HM, Shin J, Lafaille JJ, Possemato R, Pacold ME, Papagiannakopoulos TY, Kimmelman AC, Satija R, Littman DR
Cell. 2020
Groen SC, Calic I, Joly-Lopez Z, Platts AE, Choi JY, Natividad M, Dorph K, Mauck WM, Bracken B, Cabral C, Kumar A, Torres RO, Satija R, Vergara G, Henry A, Franks SJ, Purugganan MD
Nature. 2020
2019
Stuart T*, Butler A*, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R
Cell. 2019
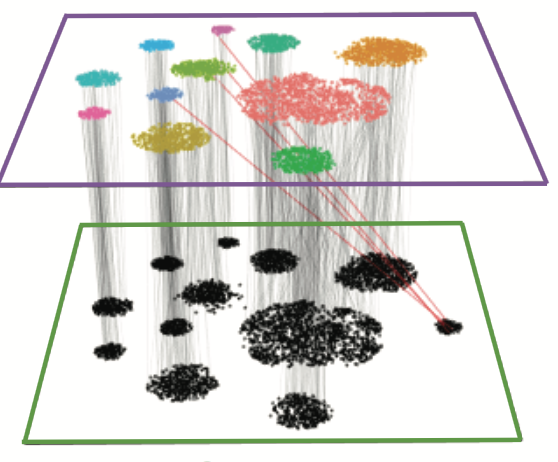
Wang W*, Niu X*, Stuart T, Estelle J, Mauck WM, Kelly R, Satija R*, Christiaen L*
Nature Cell Biology. 2019
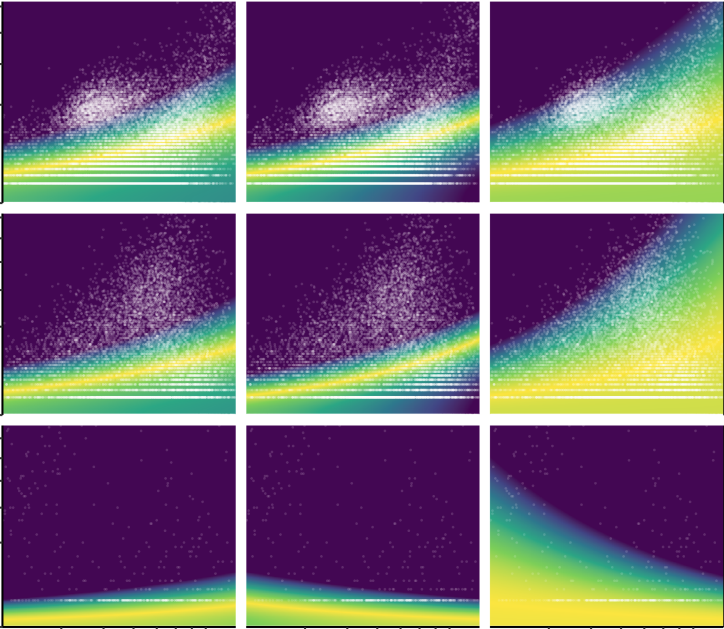
Hafemeister C, Satija R
Genome Biology. 2019
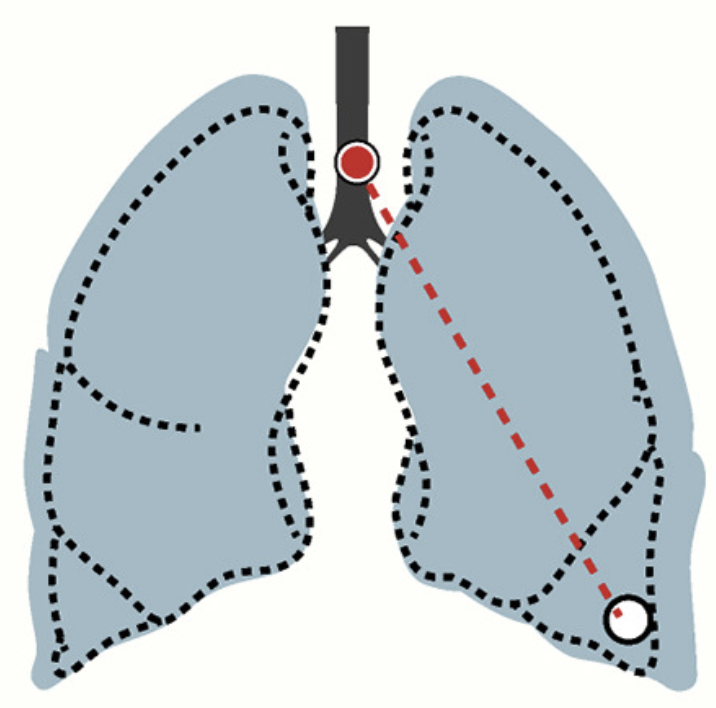
Rood J, Stuart T*, Ghazanfar S*, Biancalini T*, Fisher E, Butler A, Hupalowska A, Gaffney L, Mauck WM, Eraslan G, Marioni JC*, Regev A*, Satija R*
Cell. 2019
HuBMAP consortium
Nature. 2019

Tikhonova A, Dolgalev I, Ku H, Sivaraj K, Hoxha E, Cuesta-Dominguez A, Pinho S, Akhmetzyanova I, Gao J, Witkowski M, Guillamot MR, Gutkin M, Zhang Y, Marier C, Diefenbach C, Kousteni S, Heguy A, Zhong H, Fooksman D, Butler J, Economides A, Frenette PS, Adams R, Satija R, Tsirigos A, Aifantis I
Nature. 2019
Mimitou E, Cheng A, Montalbano A, Hao S, Stoeckius M, Legut M, Roush T, Herrera A, Papalexi E, Ouyang Z, Satija R, Sanjana N, Koralov S, Smibert P
Nature Methods. 2019
2018
Mayer C*, Hafemeister C*, Bandler RC*, Machold R, Allaway K, Jaglin X, Brito R, Butler A, Fishell G*, Satija R*
Nature. 2018
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R
Nature Biotechnology. 2018
Stoeckius M*, Zheng S*, Houck-Loomis B, Hao S, Yeung B, Mauck WM, Smibert P*, Satija R*
Genome Biology. 2018
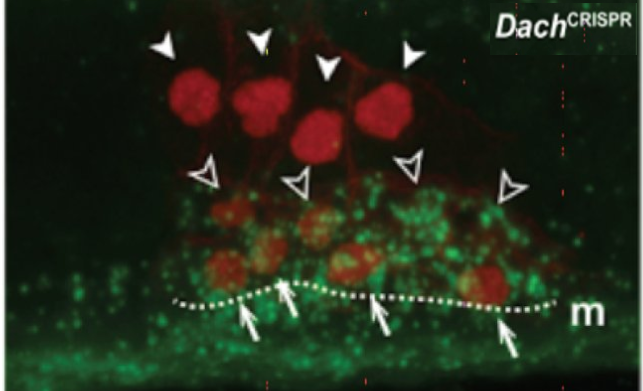
Konstantinides N, Kapuralin K, Barboza L, Satija R, Desplan C
Cell. 2018

Zheng S, Papalexi E, Butler A, Stephenson W, Satija R
Molecular Systems Biology. 2018
Stephenson W*, Donlin LT*, Butler A*, Rozo C, Bracken B, Rashidfarrokhi A, Goodman SM, Ivashkiv LB, Bykerk VP, Orange DE, Darnell RB, Swerdlow HP*, Satija R*
Nature Communications. 2018

Upadhaya S*, Sawai C*, Papalexi E, Rashidfarrokhi A, Jang G, Chattopadhyay P, Satija R, Reizis B
Journal of Experimental Medicine. 2018
2017
Papalexi E, Satija R
Nature Reviews Immunology. 2017
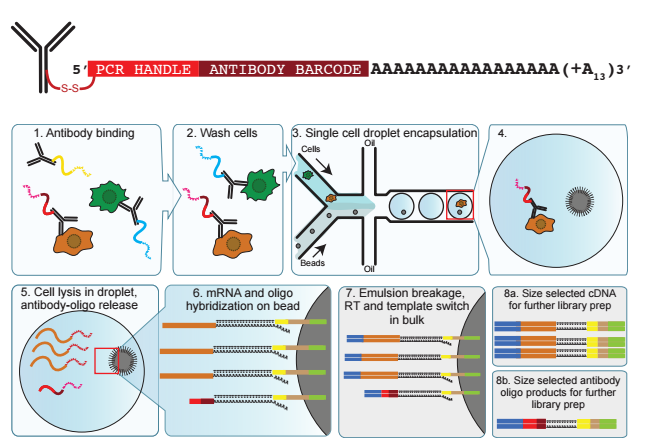
Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Swerdlow H, Satija R, Smibert P
Nature Methods. 2017
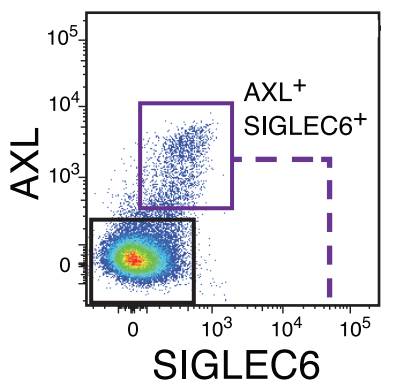
Villani AC*, Satija R*, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N
Science. 2017
Regev A, Teichmann SA, Lander ES, ... , Human Cell Atlas Meeting Participants
eLife. 2017
Gierahn TM, Wadsworth II MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK
Nature Methods. 2017

Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A, Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG, GTEx Consortium
Nature. 2017
2016
Breton G, Zheng S, Valieris R, da Silva IT, Satija R, Nussenzweig MC
Journal of Experimental Medicine. 2016
Velasco S, Ibrahim MM, Kakumanu A, Garipler G, Aydin, B, Al-Sayegh MA, Hirsekorn A, Abdul-Rahman F, Satija R, Ohler U, Mahony S, Mazzoni EO
Cell Stem Cell. 2016
Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, Neftel C, Desai N, Nyman J, Izar B, Luo CC, Francis JM, Patel AA, Onozato ML, Riggi N, Livak KJ, Gennert D, Satija R, Nahed BV, Curry WT, Martuza RL, Mylvaganam R, lafrate JA, Frosch MP, Golub TR, Rivera MN, Getz G, Rozenblatt-Rosen O, Cahill DP, Monje M, Bernstein BE, Louis DN, Regev A, Suvà ML
Nature. 2016
Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD.
Cell. 2016
2015
Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW.
Developmental Cell. 2015
Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, Kuchroo VK.
Cell. 2015

Avraham R, Haseley N, Brown D, Penaranda C, Jijon HB, Trombetta JJ, Satija R, Shalek AK, Xavier RJ, Regev A, Hung DT.
Cell. 2015

Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, Shalem O, Satija R, Raychowdhury R, Mertins P, Carr SA, Zhang F, Hacohen N, Regev A.
Cell. 2015
Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, Shalem O, Satija R, Raychowdhury R, Mertins P, Carr SA, Zhang F, Hacohen N, Regev A.
Cell. 2015
Powers A, Satija R.
Genome Biology. 2015
Satija R*, Farrell JA*, Gennert D, Schier AF, Regev A.
Nature Biotechnology. 2015

Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas A, Kamitaki N, Martersteck E, Trombetta JT, Weitz DA, Sanes JR, Shalek AK, Regev A, McCaroll SA.
Cell. 2015
Jovanovic M*, Rooney MA*, Mertins P, Przybylski D, Chevrier N, Satija R,Rodriguez EH, Fields AP, Schwartz S, Raychowdhury R, Mumbach MR, Eisenhaure T, Rabani M, Gennert D, Lu D, Delorey T, Weissman JS, Carr SA, Hacohen N, and Regev A.
Science. 2015
2014
Satija R, Shalek AK.
Trends in Immunology. 2014
Shalek AK*, Satija R*, Shuga J*, Trombetta JJ, Lu D, Gennert D, Chen P, Gertner RS, Gaublomme JT, Yosef N, Schwartz S, Fowler B, Weaver S, Wang J, Wang X, Ding R, Raychowdhury R, Friedman N, Hacohen N, Park H, May AP, and Regev A.
Nature. 2014
Trombetta JJ*, Gennert D*, Lu D *, Satija R, Shalek AK, and Regev A.
Current Protocols in Molecular Biology. 2014

Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser AJ, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, Ferrante TC, Regev A, Daley GQ & Collins JJ.
Nature. 2014
Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, & Regev A.
Cell. 2014
Lohr JG, Adalsteinsson VA, Cibulskis K, ChoudhurD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee GSM, Li T, Chabot ME, Ly A, Taplin ME, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS & Love JC.
Nature Biotechnology. 2014
2013
Shalek AK*, Satija R*, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta JJ, Gennert D, Gnirke A, Goren A, Hacohen N, Levin JZ, Park H, Regev A.
Nature. 2013
Adiconis X, Borges-Rivera D, Satija R, DeLuca DS, Busby MA, Berlin AM, Sivachenko A, Thompson DA, Wysoker A, Fennell T, Gnirke A, Pochet N, Regev A, Levin JZ.
Nature Methods. 2013
Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, Carr SA, Lander ES, Fink GR, & Regev A.
Cell. 2013
Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, Gennert D, Satija R, ShakyaA, Lu DY, Trombetta JJ, Pillai MR, Ratcliffe PJ, Coleman ML, Bix M, Tantin D, Park H, Kuchroo VK & Regev A.
Nature. 2013

2012
Satija R, Bradley RK.
Genome Research. 2012
Joyce PI, Satija R, Chen M, Kuwabara PE
PLoS Genetics. 2012
2010
Satija R, Hein J, Lunter GA.
Bioinformatics. 2010